Figures & data
Figure 1. Adhesion of K. pneumoniae to human retinal epithelial cells. (A) Adhesion to ARPE-19 cells by PLA-endophthalmitis-associated K. pneumoniae clinical isolates (as indicated). NTUH-A4528 was used as a K2 type control strain. The adhesion rate was expressed as the proportion of the inoculum that adhered (∼1 × 10−4 of the inoculum). Data are presented as the mean ± SEM from 3 independent trials. (B) ARPE-19 adhesion of the K. pneumoniae NTUH-K2044 wild-type and its isogenic mutants (as indicated). The adhesion rate was expressed as the proportion of the NTUH-K2044 wild-type strain that adhered. Data are presented as the mean ± SEM from 3 independent trials. *, P < 0.05; **, P < 0.01. (C) Mapping of the transcription start site for the yfgL gene. The promoter region and the proposed −10 and −35 regions are indicated by gray shading. The transcription start site is indicated by +1 with arrowhead. The primers shown here indicate the relative positions in construction of the yfgL complementation strain. (D) Growth experiments with the wild type, yfgL, clpS and fim mutant strains in LB broth or MM. Overnight cultures of the NTUH-K2044 wild-type, yfgL, clpS, and fim mutant strains were inoculated (separately) into fresh LB medium or MM and grown at 37°C, respectively. The growth of bacteria was monitored hourly by plating of serial dilutions on LB agar and counting of CFUs following overnight growth of the plates. Data are presented as the mean ± SEM from 3 independent trials.
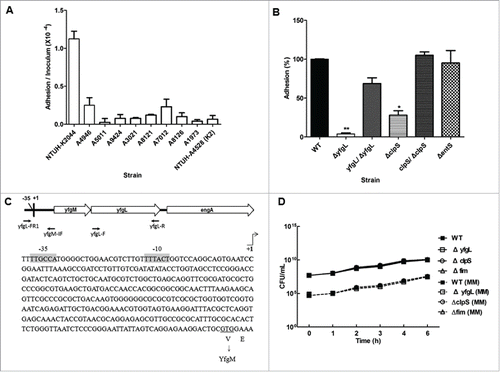
Table 1. The K. pneumoniae NTUH-K2044 hyperadherent clones.
Figure 2. The effects of YfgL on K. pneumoniae adherence. (A) Specificity of rabbit polyclonal antibodies raised against YfgL. Total outer membrane protein extracts from normalized bacterial suspensions (4 × 109 CFU) of the NTUH-K2044 wild-type, ompA, yfgL mutant, and yfgL complementation strains grown in LB were analyzed by 12% SDS-PAGE, and stained with Coomassie brilliant blue (left), respectively. ompA deletion strain was used as a control strain. For western blots, proteins were probed with polyclonal antibodies raised against YfgL (1:10,000). The positions of OmpA and YfgL are indicated. (B) The NTUH-K2044 was incubated with rabbit polyclonal anti-K1 antibodies (as a positive control) or with rabbit polyclonal anti-YfgL antibodies, respectively. After incubation, Alexa Fluor 488 anti-rabbit IgG were added for staining, and then the fluorescence was observed. Scale bar, 10 μm. (C) Adhesion of the K. pneumoniae NTUH-K2044 wild-type, yfgL, and fim mutant strains (as indicated) to ARPE-19, Caco-2, or A549 cells. The adhesion rate was expressed as the proportion of the NTUH-K2044 wild-type strain that adhered. Data are presented as the mean ± SEM from 3 independent trials. *, P < 0.05; **, P < 0.01. (D) Immunofluorescence image analysis showing the adhesion of ARPE-19 cells by the NTUH-K2044 and the yfgL mutant. Cultured cells were infected by a high MOI of the NTUH-K2044 or the yfgL mutant for 15 min. Adherence K. pneumoniae (green) were incubated with rabbit anti-K1 polyclonal antibodies (1:1000) and then stained with Alexa Fluor 488 anti-rabbit IgG (Invitrogen). The actin cytoskeleton and DNA were stained with rhodamine-phalloidin (red) and DAPI (blue). Scale bar, 8 μm.
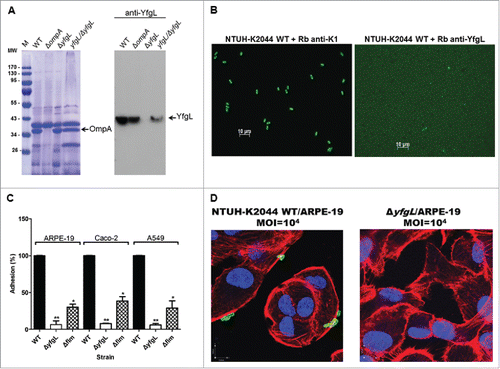
Table 2. Effect of yfgL deletion on transcription of genes in the K. pneumoniae NTUH-K2044 by microarray.
Figure 3. The roles of YfgL in type-1 fimbriae production and cell invasion of K. pneumoniae. (A) Orientation of the fim phase-switch element in the K. pneumoniae NTUH-K2044 strains following overnight culture in either static or shaking LB broth. Depending on the orientation of the fim invertible element, this method generates paired fragments of different sizes (613 and 404 bp when in the “on” orientation, 840 and 177 bp when in the “off” orientation). Lane M contains DNA molecular size markers. (B) Effect of mutations in the yfgL or fim genes on yfgL and fim transcription in the wild-type, yfgL, fim mutants and yfgL complementation strains. Quantitative real-time expression of the yfgL and fim genes in response to LB broth shaking cultures in the wild-type strain, yfgL, fim mutants and yfgL complementation strains. The data represent averages from 3 independent trials, and the error bars represent the SEM. (C) Three-dimensional image analysis showing the invasion of ARPE-19 cells by K. pneumoniae NTUH-K2044. Cultured cells were infected by a high MOI of the NTUH-K2044 carrying TA-GFP (encoding greenfluorescent protein; green) for 2 h. The actin cytoskeleton and DNA were stained with rhodamine-phalloidin (red) and DAPI (blue). Images from confocal microscopy with z-stacking were analyzed by XY, XZ, and YZ sections. Intracellular K. pneumoniae are indicated by white arrows. Scale bar, 8 μm. (D) Invasion of ARPE-19 cells by NTUH-K2044 wild-type, yfgL mutant, and yfgL complemented strains. The invasion rate was expressed as the proportion of the NTUH-K2044 wild-type strain that invaded. Data are presented as the mean ± SEM from 3 independent trials. **, P < 0.01. (E) Effects of yfgL deletion on outer membrane protein level of K. pneumoniae. Total outer membrane protein extracts from normalized bacterial suspensions (4 × 109 CFU) of K. pneumoniae NTUH-K2044 wild-type, ompA, yfgL mutant, and yfgL complementation strains grown in LB or NB broth were analyzed by 12% SDS-PAGE, and stained with Coomassie brilliant blue, respectively. ompA deletion strain was used as a control strain. The positions of OmpA and OmpK35/OmpK36 are indicated.
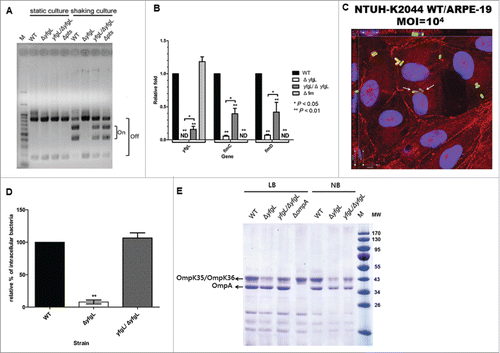
Table 3. Characteristics of the NTUH-K2044 wild-type and its derivatives.
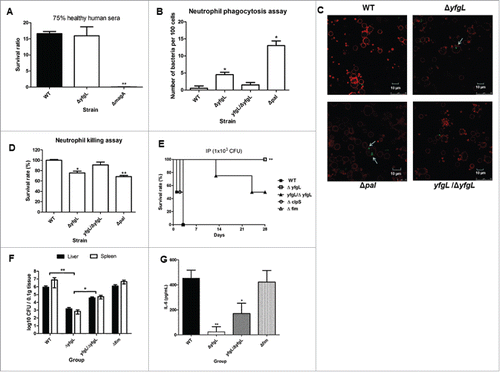
Figure 4. The roles of YfgL in virulence during K. pneumoniae infection. (A) Serum sensitivity assays of resistance to killing by nonimmune healthy human serum of the NTUH-K2044 wild-type and the yfgL-gene deletion mutant strains. The data represent the means of 3 independent trials; the error bars represent the standard deviations. A mean survival ratio ≥1 corresponds to serum resistance. ** P <0.01 by Student's t test (compared to the wild-type strain). The magA-gene deletion mutant is a CPS-deficient strain of NTUH-K2044 and used as a control. ((B)and C) Determination of phagocytosis resistance of the wild type, the yfgL-gene mutant and the yfgL complementation strains by human neutrophil. The pal-gene deletion strain is sensitive to human neutrophil phagocytosis and killing and used as a control. Bacteria carrying the GFP plasmid were incubated with human neutrophils for 45 min and observed under a confocal microscope. Bacteria phagocytosed by human neutrophils were counted under a confocal microscope. Data are presented as mean ± SEM of 3 independent trials. ** P < 0.01 or * P < 0.05 by Student's t test (compared to the wild-type strain). A representative confocal section through the middle of the cell was shown for observation of intracellular bacteria. Arrows denote intracellular bacteria. Scale bar, 10 μm. (D) Bacterial susceptibilities to killing by human neutrophils of the wild type, the yfgL-gene mutant and the yfgL complementation strains after 45 min of incubation are presented. Survival rate indicates percent survival of wild-type or mutant strains calculated on the basis of viable counts relative to those for the no-neutrophil controls. Data are presented as mean ± SEM of 3 independent trials. ** P < 0.01 or * P < 0.05 by Student's t test (compared to the wild-type strain). (E) BALB/c mice (4 mice per group) were infected with the NTUH-K2044 wild-type and isogenic mutant strains at an IP dose of 1 × 103 CFU/animal. Survival of mice was monitored for 4 weeks. ▪, NTUH-K2044; □, yfgL mutant (yfgL vs. parent, **, P = 0.008; log-rank test); ○, clpS mutant; △, fim mutant; ▴, yfgL complementation strain. ((F)and G) BALB/c mice (4 mice per group) were inoculated by IP injection with equivalent doses (1 × 103 CFU) of the NTUH-K2044 wild-type, yfgL mutant, or fim mutant strains. Surviving animals were sacrificed at 20 hours after challenge. Bacterial loads were measured in the liver and spleen (F); IL-6 levels were measured in the serum (G). Tissue counts (log10 CFU) were standardized per 0.1 g wet organ weight. IL-6 levels were measured by ELISA. Data are presented as mean ± SEM. **, P < 0.01 by Student's t-test (compared to the wild-type strain); other comparisons were not statistically significant (P ≥ 0.05).