Figures & data
Figure 1. A C-terminal toxin domain carried by an extended Hcp exhibited antibacterial activity in E. coli. (A) Schematic diagram of the genetic organization of the T6SS locus and Hcp-ET1 effector-immunity pair in strain STEC004. The related sequences have been submitted to GenBank under the accession nos. KX118291 and KX118294, and exhibit high sequence identity to that of sequenced O104:H7 strain C227–11. (B) Growth competition assays between indicated E. coli donor and recipient strains. Experiments were initiated by mixing the donor and recipient bacteria at a CFU ratio of 5:1. The Log10 CFU values of the surviving recipient cells are shown on the y-axis. The pGEN MCS with the target gene inserted under the control of Pcm promoter was used as the complementary plasmid. Asterisks indicate significant differences in the outcome of competition assay for the same donor strain against the indicated recipient strains (**, P < 0.05). Error bars indicate standard deviations (SDs) of 3 independent experiments. (C) Hcp-ET1 degraded plasmid-DNA pUC19 completely in vitro. Purified Hcp-ET1 was incubated with linear pUC19 DNA in the presence and absence of cognate ETI1 or non-cognate Crp (cAMP receptor protein). Crp is a stable protein that exists generally in the cytoplasm of E. coli, and was used as a control for cytoplasmic toxin.Citation60 Reactions were analyzed by native agarose-gel electrophoresis and Goldview (Vazyme, China) staining. All samples were also analyzed by SDS-PAGE and Coomassie blue staining to confirm the proteins' stabilities.
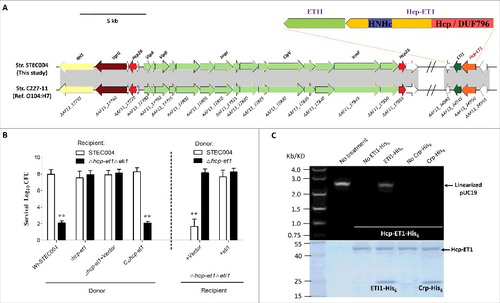
Figure 2. Hcp-ET1 is a T6S-dependent effector that degrades target cell DNA. (A) The toxicity of Hcp-ET1 was dependent on the DUF796 domain and a functional T6SS. The donor and recipient strains were mixed together at a ratio of 3:1, and then incubated for 6 h at 30 °C. The bacterial plaques (NalR recipients) from left to right were increasing serial 10-fold dilutions on NalR eosin-methylene blue agar. The pGEN MCS with the hcp-et1 gene inserted under the control of Pcm promoter was used for complementation (strain CΔhcp-et1). (B) The Hcp-ET1 degraded the plasmid DNA of target cells. The recipient (Δhcp-et1Δeti1 with pUC19) was mixed with the indicated donor strains at a ratio of 3:1, and then collected after a 6 h incubation at 30°C. Equal cell mass was collected from each group, and plasmid DNA was extracted. The linear plasmid DNA from each preparation was analyzed by agarose-gel electrophoresis and Goldview (Vazyme, China) staining. The “untreated STEC004” means only STEC004 cells that were not mixed with recipient cells for the bacterial competition assay. (C) Effects of Hcp proteins on Hcp-ET1 delivery. Donor cells were STEC004 and its derivative hcp and T6SS mutants, as indicated. Experiments were initiated by mixing the donor and recipient bacteria at a CFU ratio of 5:1. The Log10 CFU values of the surviving recipient cells are shown on the y-axis. Error bars indicate standard deviations (SDs) of 3 independent experiments. (D) Fluorescence microscopy for E. coli STEC004 co-incubation. The GFP-labeled donor cells (containing plasmid pGEN-pcm-GFP) were co-incubated with the unlabeled target cells lacking eti1 on an LB plate at a 1:1 ratio. Samples from 0 and 12 h were stained with DAPI to visualize genomic DNA. White arrows indicated anucleate target cells. GFP, green fluorescent protein.
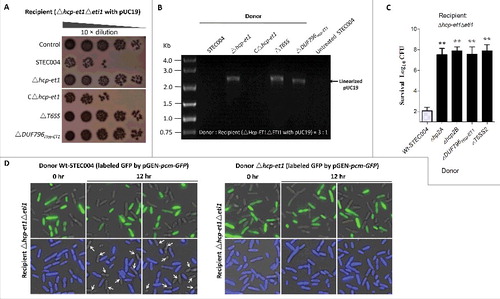
Figure 3. The extended Hcps harbored diverse ET-toxins in multiple bacterial species from Enterobacteriaceae. The genomic organization of the Hcp-ET modules was shown. Red boxes indicate the DUF796 domains. The dark-blue and brown boxes indicate the toxic domains, and the sky-blue boxes indicate the putative immunity genes. All of the predicted Hcp-ET effector-immunity pairs are listed in Table S1, and some representative Hcp-ET pairs from 5 clans are presented here.
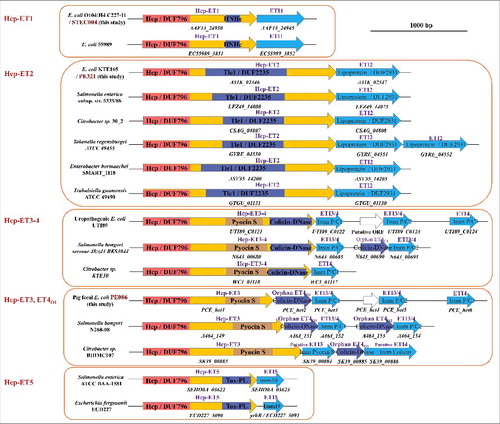
Figure 4. Hcp-ET2 functions as Tle1 family phospholipase for interbacterial competition. The sequence of the Hcp-ET2 effector-immunity pair from ETEC PE321 has been submitted to GenBank under the accession no. KX118290. (A) Hcp-ET2 is a T6S-dependent Tle1 phospholipase. The phylogenetic tree of ET2 and Tle1 phospholipase effectors was constructed based on an alignment of catalytic motifs, as described previously.Citation14 ET2 proteins (labeled red) were clustered into the same branch with 2 known Tle1 members of E. coli KD1 and Salmonella enterica ATCC BAA-1581. Sequence logos analysis indicated that ET2 proteins also contained a GxSxG catalytic motif, which has been identified as the conserved catalytic motif of Tle1–4 families.Citation14 (B) Hcp-ET2 provided a competitive growth advantage under cell contact-promoting conditions. Donor and recipient strains were mixed at a 5:1 ratio, incubated for 6 h on solid media and differentiated using blue/white screening. The pGEN MCS with the hcp-et2 gene inserted under the control of Pcm promoter was used for complementation (CΔhcp-et2). The Log10 CFU values of the surviving recipient cells are shown on the y-axis. Asterisks denote significant difference in the outcome of the competition assay for the same donor strain against the indicated recipient strains (**, P < 0.05). Error bars represent the SDs for 3 independent experiments. (C) ETI2 could bind to its cognate Hcp-ET2 directly. Purified Hcp-ET2 and ETI2-His6 proteins were mixed at equimolar ratios, and then purified by Ni2+-affinity chromatography. Crp (cAMP receptor protein) is a stable protein in the cytoplasm of E. coli generally, and was used as a control in this assay. Input samples represent the protein mixtures before chromatography. All fractions were analyzed by SDS-PAGE and Coomassie blue staining.
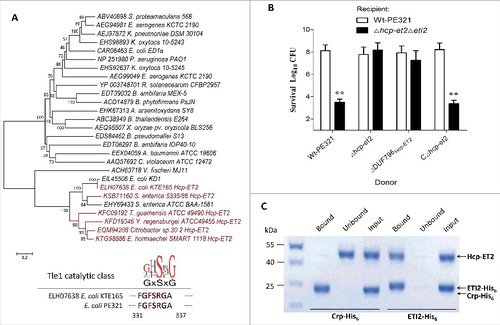
Figure 5. The ET3 domain is involved in interbacterial antagonism. The sequence of the Hcp-ET3_ET4O1 module from ETEC PE086 has been submitted to GenBank under the accession number KX118289. (A) Artemis Comparison Tool (ACT) comparison of the Hcp-ET3–4 module from uropathogenic E. coli UTI89 and the Hcp-ET3_ET4O1 module from ETEC PE086. The PEC_het4–6 is located downstream of PEC_het1–3 in the genome of PE086. The highly conserved genomic region (red blocks denote nucleotide conservation) included these 2 loci from 5’ to 3’, and showed 3 duplications of the et4-eti3/4 fragment. (B, C) Hcp-ET3 provided a competitive growth advantage under cell contact-promoting conditions between the indicated E. coli donor and recipient strains. Experiments were initiated by mixing the donor and recipient bacteria at a CFU ratio of 5:1. The Log10 CFU values of the surviving recipient cells are shown on the y-axis. Asterisks denote significant difference in the outcome of the competition assay of the same donor strain against the indicated recipient strains (**, P < 0.05). Error bars represent the SDs for 3 independent experiments. (D) Growth curves of E. coli cells producing Hcp-ET3 or co-producing Hcp-ET3 and ETI3/4. The cytoplasmic expression of et3 and eti3/4 was induced by L-Arabinose at the indicated time (arrow). A native ribosome binding sites of E. coli located upstream of eti3 gene contributed to achieve its co-expression (blue curve). The experiments were run in triplicate.
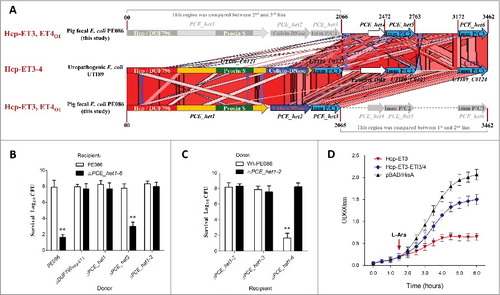
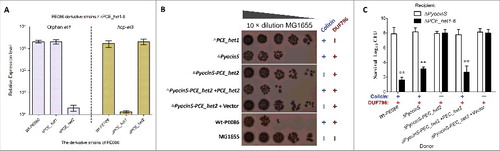
Figure 6. Orphan ET4 toxin is functional for interbacterial competition. (A) The orphan et4 might be co-transcribed with the hcp-et3 by the same promoter. The data were normalized to the transcription level of the housekeeping gene tus. The relative expression levels represent the mean ± SD for 3 independently isolated RNA samples. (B) The toxic activity of ET4O1 was dependent on the retained DUF796 domain and functional T6SS. The recipient (screened NalR MG1655, a reference strain for T6SS killingCitation40) and indicated donor strains were mixed together at a ratio of 3:1, and then incubated for 6 h at 30°C. The bacterial plaques from left to right were increasing serial 10-fold dilutions on NalR eosin-methylene blue agar. The pGEN MCS with the target gene inserted under the control of the Pcm promoter was used as the complementary plasmid. The presence and absence of Colicin-DNase (ET4) in donor strains were indicated by blue “+” and “−,” respectively. Similarly, red icons indicate the presence and absence of DUF796. (C) Growth competition assays between the indicated donor and recipient strains. Experiments were initiated by mixing the donor and recipient bacteria at a CFU ratio of 5:1. The Log10 CFU values of the survivaing recipient cells are shown on the y-axis. Asterisks denote significant difference in the outcome of the competition assay for the same donor strain against the indicated recipient strains (**, P < 0.05). Error bars represent the SDs for 3 independent experiments.