Figures & data
Figure 1. Identification of SCIN as target of humAb 6D4. Western blot analysis using humAb 6D4 on proteins from cell pellet (P) and growth medium fractions (supernatant; S) of the S. aureus (Sa) strains NCTC8325 and NCTC8325 ΔspaΔsbi (A), and the growth medium fractions of strains NCTC8325 ΔpknB and NCTC8325 ΔpknB ΔΦ13 (B). Western blot analysis of the growth medium fractions of L. lactis pNG4210::scn or pNG4210::chips secreting the SCIN or CHIPS proteins, respectively, using anti-His-tag antibodies (C), or humAb 6D4 (D). Molecular weights (kDa) of marker proteins are indicated next to panel A.
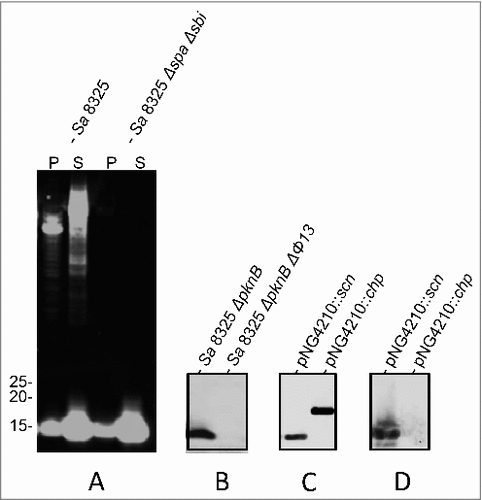
Figure 2. HumAb 6D4 binds to the C-terminal part of the first α-helix of SCIN. Proteins from E. coli Rosetta Gami expressing SCIN-OrfD chimera were separated using SDS-PAGE. The expressed chimera of SCIN and OrfD are schematically presented (A). The 3 helices (α1, α2 and α3) and the active site region of SCIN (in gray shading) are indicated. SCIN residues (gray) were exchanged with corresponding residues from OrfD (black). Exchanged residues (in parentheses) are: CH-N (1–13), CH-C (83–85), CH-α1N (1–25), CH-α1C (26–36), CH-α2N (37–48), CH-α2C (49–58), CH-α3N (59–72), CH-α3C (73–86), CH-α1CA (26–30), CH-α1CB (31–36), CH-α2NA (37–42), and CH-α2NB (43–48). Gels were stained with simply blue to verify protein production (B), and the produced proteins were specifically detected by immunoblotting with an anti-His-antibody (C) or the humAb 6D4–800CW (D). The positions of molecular weight marker proteins (kDa) are shown next to the gel and Western blot images.
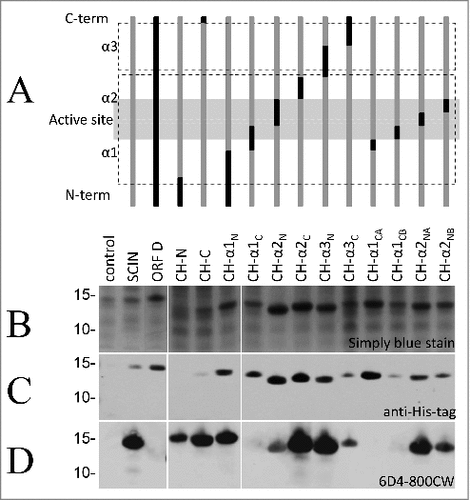
Figure 3. Binding of the humAb 6D4 to SCIN produced by different laboratory strains and clinical isolates of S. aureus. Western blotting analysis using humAb 6D4–800CW to detect SCIN in the cell pellet (P) or growth medium (S) fractions of S. hominis, S. haemolyticus and the S. aureus strains Newman, USA300, Mu50, MW2, N315, COL, NCTC8325–4, MRSA252 and MSSA476 (A), or in the growth medium fractions (supernatant) of 24 clinical S. aureus isolates named A-J and L-Y (B). Molecular weights (kDa) of marker proteins are indicated to the left of panels A and B. Loading of comparable amounts of proteins was confirmed by Simply Blue staining (not shown).
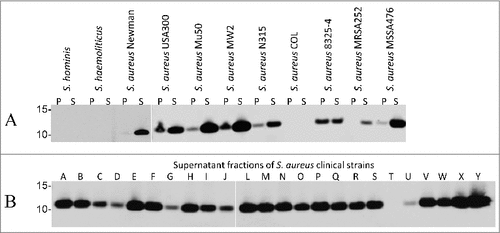
Figure 4. Binding of SCIN to S. aureus cells increases upon incubation in serum. Western blotting analysis of S. aureus Newman ΔspaΔsbi cells collected by centrifugation (P) and growth medium fractions (S) using 6D4–800CW. The presence or absence of C3 convertases due to serum incubation, and the addition or absence of SCIN are indicated with + or -, respectively.
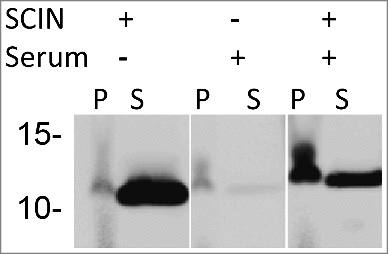
Figure 5. Binding of the humAb 6D4 to whole cells of S. aureus. Plates were coated with whole cells of various S. aureus clinical isolates or laboratory strains harvested from cultures in the mid-exponential growth phase where the growth medium was supplemented with human serum. 6D4–800CW was used for the detection of cell-bound SCIN, and an α-his-tag antibody was used as a negative control. Fluorescence readings at 800 nm are plotted relative to the binding of 6D4–800CW to S. aureus Newman ΔspaΔsbi. All measurements were performed in triplicate and the mean ± standard error (error bars) is shown. (A) concentration-dependent binding of 6D4–800CW to S. aureus Newman ΔspaΔsbi, Newman wild-type (wt), the clinical S. aureus isolate P, or the MRSA strain USA300 is indicated in in black symbols; the lack of binding of the α-his-tag control antibody to S. aureus Newman ΔspaΔsbi, Newman wild-type (wt), isolate P, or USA300 is shown in gray symbols. (B) Binding of 6D4–800CW to S. aureus Newman ΔspaΔsbi, various clinical S. aureus isolates and the sequenced S. aureus strains USA300, Mu50, MW2, N315, COL, 8325–4, MRSA252, MSSA476, Newman wild-type (WT) and Newman ΔspaΔsbi is indicated with black bars; binding of 100 ng/mL isotype control antibody IQNPA to the S. aureus clinical isolates and sequenced S. aureus strains as specified is indicated with white bars.
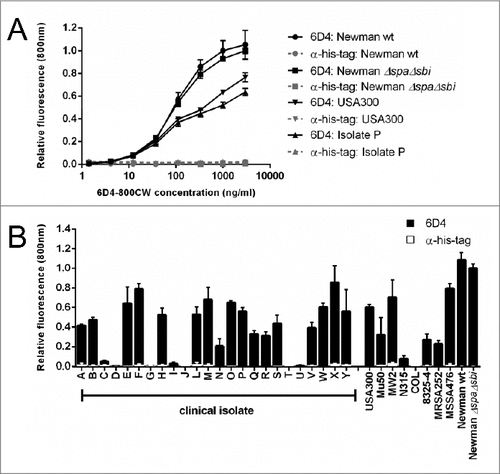
Figure 6. Serum-incubated S. aureus cells display elevated levels of SCIN binding. Phase contrast (panels A, C, E) and subsequent fluorescence microscopy at 800 nm (panels B, D, F) of cells of S. aureus Newman ΔspaΔsbi collected from an overnight culture. Specifically, the panels show cells from the overnight culture (A, B), cells treated with serum but without the addition of SCIN (C, D), and cells treated with serum and added SCIN (E, F). Cell-bound SCIN was detected using 6D4–800CW.
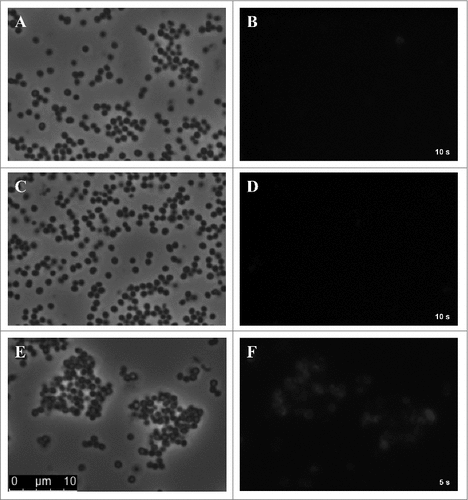
Figure 7. Impact of humAb 6D4 on SCIN activity. (A) C3b deposition on S. aureus Newman ΔspaΔsbi cells upon preincubation of SCIN with humAb 6D4 (■). C3b deposition was monitored by flow cytometry. As a negative control, SCIN was preincubated with buffer (♦), or control IgG (▴). Each data point represents the mean ± standard error (error bars) of 3 independent experiments. (B) Reduced SCIN-mediated protection of rabbit erythrocytes against lysis by complement upon incubation of SCIN with humAb 6D4 (■). Hemolysis was quantified by pelleting of erythrocytes and subsequent measurement of the absorbance of supernatants at 450 nm. As a control, SCIN was preincubated with buffer (♦), or control IgG (▴). Each data point represents the mean ± standard error (error bars) of 2 separate experiments.
Table 1. Strains and plasmids used in this study.
Table 2. Primers used for detection or cloning of scn and chp genes.
