Figures & data
Figure 1. The 3D structures of bacterial cytokine-binding proteins colored from the N-terminus (blue) to the C-terminus (red). (A) The N-terminal PapC domain of Y. pestis Caf1A (PDB:4BOE), the β, N0 and N1 domains of N. meningitidis PilQ (PDB:4AV2), the N-terminal transmembrane domain (PDB:4RLC), and the C-terminal domain (PDB:5U1H) of P. aeruginosa OprF are located in the outer membrane of gram-negative species. (B) E. coli IrmA (PDB:5EK5) and N. meningitidis PilE (PDB:5JW8) are secreted to and face the extracellular space, respectively. The figures were prepared with PyMol (www.pymol.org)
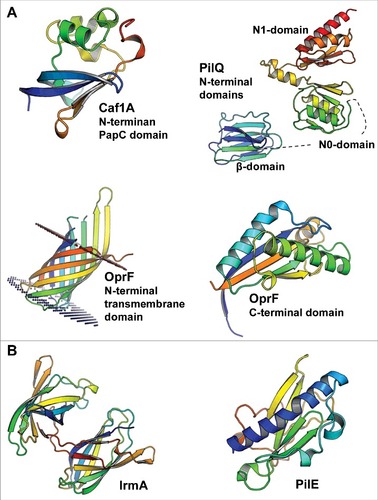
Figure 2. Structural similarities of bacterial cytokine-binding proteins and proteins involved in cytokine signaling. The bacterial proteins are shown in blue. (A) IrmA compared with the N-terminal Ig-fold domain of β-domain of IL-2R (PDB:2B5I), (B) the C-terminal domain of OprF compared with macrophage migration inhibitory factor (MIF) (PDB:4P7M), (C) the N-terminal PapC domain of Caf1A compared with transcription elongation factor b polypeptide 2 (TCEB2) (PDB:2IZV), and (D) the N0 domain of PilQ compared with human granulocyte macrophage colony stimulating factor receptor (GM-CSF) (PDB:5D71) are shown. Superimpositions were performed using the secondary-structure matching (SSM) tool in Coot.Citation59 The figures were prepared with PyMol (www.pymol.org)
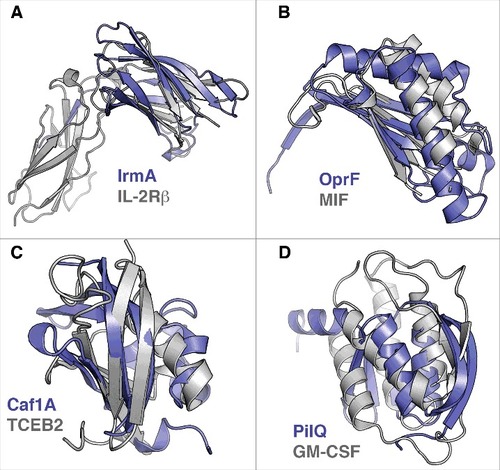
Table 1. Cytokine-binding bacterial proteins, their ligands and their virulence properties
