Figures & data
Table 1. Vibrio parahaemolyticus strains used in this study.
Figure 1. Survival of G. mellonella following infection with 100 CFU per larvae of V. parahaemolyticus strains RIMD 2210633, G35, T08, PSU 3565, T024, T03, V05/070 or V13/003A. The results shown are the means of three experiments, each using groups of 10 larvae per strain. The error bars indicate standard deviation.
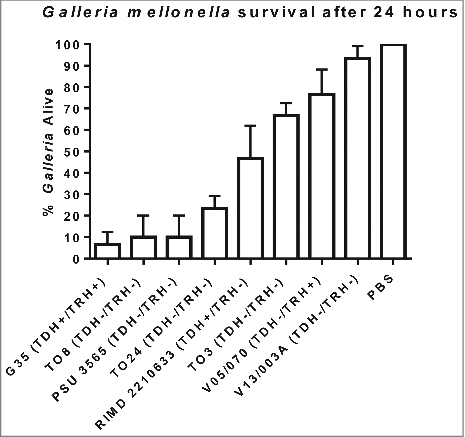
Figure 2. Survival of G. mellonella after 48h following challenge with 1–10 CFU per larvae of V. parahaemolyticus T24 or T24:ΔmutT. Groups of 10 larvae were challenged. The results shown are the means of three replicates. The error bar indicates standard deviation.
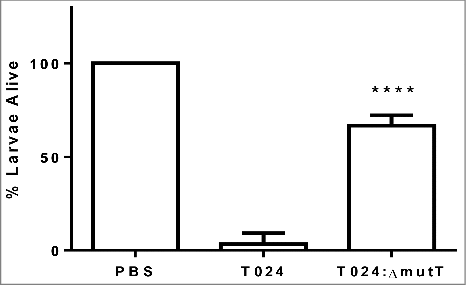
Figure 3. Growth of T024 or T024:ΔmutT at 30°C in minimal media measured by optical density (left panel) or CFU (right panel). At 8 hr there is significant difference between the wildtype and the T024:ΔmutT (determined by a paired T test P < 0.05) while no significant difference is seen with the cell counts.
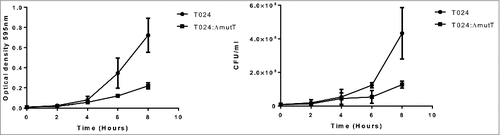
Figure 4. A phylogenetic tree compiled from recA, gyrB, pyrH and atpA sequences from sequenced V. parahaemolyticus strains including the reference strain RIMD 2210633.
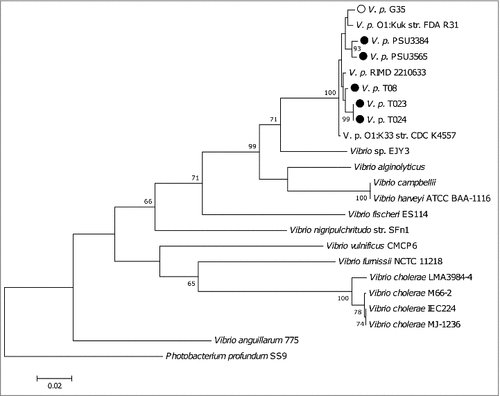
Table 2. Oligonucleotide primers used in this study.
Table 3. Presence or absence of chromosomal regions in 5 non-toxigenic strains (T08, T023, T024, PSU 3565 and PSU 3384) and 1 toxigenic strain (G35) when compared to reference strain RIMD2210633 and their corresponding absence/presence in the 6 genome-sequenced strains.
