Figures & data
Figure 1. Possible nuclear and mitochondrial genetic genotypes in Cryptococcus species in progeny from crosses contributing to their virulence. 1a) In-group cross (within species), two possible scenarios: (ai) same sex mating (α-α), recombinant mitochondrial genomes, with isogenic nuclei, and (aii) opposite sex mating (a-α) with both recombinant mitochondrial and nuclear genomes generated in progeny. In these crosses mitochondrial inheritance is biparental, with contribution from single parent at a time. The exception is crosses between isogenic strains, where mitochondrial inheritance is uniparental. 1b) Outgroup cross (interspecies): the nuclear genome can be genetically incompatible leading to post-zygotic meiotic failure, but retaining the possibility of recombination amongst mitochondrial genomes. In such a scenario progeny will have recombinant mitochondria with nuclei isogenic to one of the parents. 1c) Possible mitochondrial and nuclear genome combinations: (i) recombinant mitochondrial genome in isogenic nuclear background, (ii) recombinant mitochondrial genome in a recombinant nuclear background, (iii) non-recombinant mitochondria from first parent with non-recombinant nuclei from another parent and (iv) vice versa of iii. Interorganelle (nuclear-mitochondrial) regulatory loops on each other are shown. Light blue circles with green outline and dark blue circles with red outline represents nuclei with different genetic background respectively. Blue circles with mixed dotted outline represents recombinant nuclei. The a and α represents mating types. Ellipse with green and red outline represents mitochondria with different genetic background respectively. Ellipse with red green dotted outline and yellow color represents recombinant mitochondria. Curved arrows in 1c represents interorganelle regulatory signaling loops for nucleus to mitochondria (purple) and mitochondria to nucleus (brown).
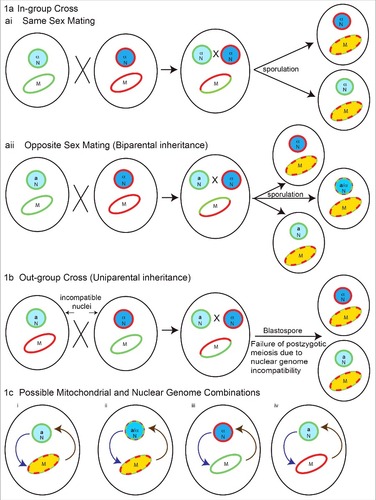
Figure 2. Overview of mitochondrial crosstalk with other organelles and cellular components such as the nucleus, endoplasmic reticulum (ER), the sorting and assembly machinery (SAM), plasma membrane (PM), and cell wall. 2a) Mitochondrial dysfunction or defects triggers mitochondrial-nuclear retrograde signaling. Loss of mitochondrial genome and mitochondrial fusion defect via unknown mechanisms activates nuclear retrograde (RTG) genes. 2ai) The mitochondria-nuclear retrograde signaling genes have role in regulating expression of PDR genes. Change in RTG gene expression in turn affect the expression of PDR3. Pdr3/CgPdr1 has an auto-regulatory loop (dotted curved arrow inside nucleus) and RTG functions to regulate Pdr3 expression. Pdr3/CgPdr1 and other unknown nuclear factors regulates expression of drug transporter Pdr5/CDR1/CDR2 by acting on its PDRE. One of the factors is Lge1, a nuclear protein downstream of Psd1, as ρ° cells increase the expression of Pdr3 and Pdr5. 2aii) Increase in the expression of drug transporter Pdr5 leads to increase in drug efflux hence drug resistance. 2aiii) Mutation and dysfunction of mitochondrial respiratory complex perturbs overall cells energy state and also possibly affects the membrane fluidity (perturbed ergosterol biosynthesis), which in turn leads to down regulation of drug transporters CDR1/CDR2 and hence increased drug susceptibility. 2aiv) Mitochondrial dysfunction, such as a fusion defect or loss of the mitochondrial genomes affects lipid biosynthesis and can in turn perturb the membrane permeability. 2av) Drug transporters Cdr1/Cdr2/Pdr5 have a role in synthesis of phospholipids, their distribution and transport across the plasma membrane. 2b) Mitochondria are the site for heme biosynthesis and at the center of regulating cellular heme homeostasis and its link to azole drug resistance. (2bi-2biii) Heme is the cofactor for enzymes involved in sterol biosynthesis enzymes, as a cofactor for mitochondrially-located respiratory complex enzymes and ER-localized cytochrome P450. 2biv) The heme-binding catalytic site for cytP450 can also be bound by azole drugs, and hence when targeted can impair ergosterol biosynthesis. 2bv) Heme-binding protein, Dap1, co-localizes with cytP450 at the ER. 2bvi) Dap1 dependent stabilization of cytP450, possibly by channeling of electrons from Dap1 heme to cytP450 heme. 2c) Role of mitochondrial outer membrane complex SAM (sorting and assembly machinery) in maintaining cell wall integrity and mitochondrial outer membrane function. 2ci) Dysfunction of SAM components impact mitochondrial membrane assembly and sorting of proteins through mitochondrial membranes and matrix. 2cii) Mutations or dysfunction of SAM core components leads to phospholipid synthesis problems which in turn affects the distribution of glycosylphosphatidylinositol (GPI) anchor protein and lead to a reduction in cell wall integrity. 2d) Involvement of ERMES (endoplasmic reticulum-mitochondria encounter structure) and the mitochondrial-ER contact site in maintaining mitochondrial morphology. ERMES mutants have perturbed cell wall remodeling with decreased expression of cell wall glycosidase PHR1 and 1,3-β-glucan affecting cell wall integrity.
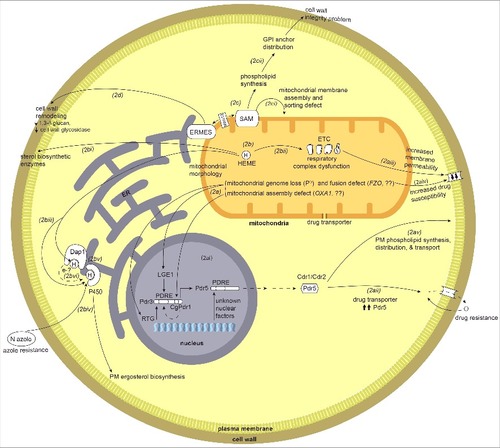
Table 1. Predicted mitochondrial proteins of Candida albicans that are involved in virulence. Genes listed in the Candida Genome Database [Citation150] (http://www.candidagenome.org/cgi-bin/phenotype/phenotype.pl?observable = virulence) with a role in virulence of C. albicans were used for prediction of mitochondrial localization. Genes encoding proteins with localization scores ≥ 50% by MitoProt II-v1.101 and/or PSORT II are listed.
