Figures & data
Figure 1. Distinction of PPAD sorting types I and II P. gingivalis isolates were cultured for four days in BHI medium. Subsequently, bacterial cells were separated from the growth medium, and growth medium fractions, containing OMVs, were used for immunoblotting with PPAD-specific antibodies. (A) P. gingivalis reference strain W83 and the isogenic PPAD deletion mutant. (B) P. gingivalis clinical isolates. Names of sorting type II isolates are underlined. Molecular weights of marker proteins and different PPAD species are indicated.
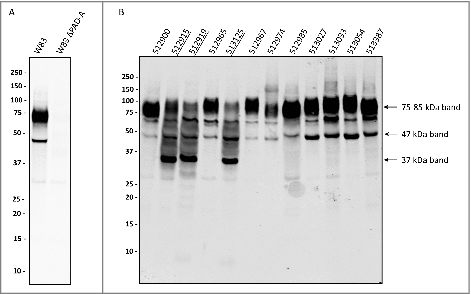
Figure 2. Association of PPAD species with OMVs Growth medium fractions (designated ‘supernatant’) of P. gingivalis sorting type I and II isolates were subject to ultracentrifugation. Subsequently, the supernatant and pellet fractions were analyzed by immunoblotting as indicated for . Samples relating to the reference strain W83 and ATCC 33277, the respective PPAD deletion mutants, and sorting type I and II isolates are indicated with names of type II isolates underlined. Molecular weights of marker proteins and different PPAD species are indicated.
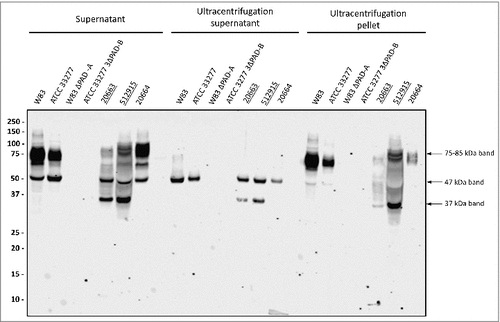
Figure 3. OMV formation by sorting type I and II isolates of P. gingivalis Electron micrographs of vesiculating cells of (A) the P. gingivalis type strain W83, and (B) the sorting type II isolate MDS33. Electron micrographs of OMVs collected from (C) strain W83, (D) the sorting type I isolate 505700, and the sorting type II isolates (E) 505759 and (F) 512915.
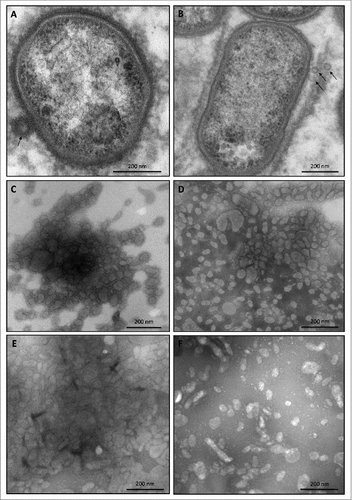
Figure 4. Protease inhibitors suppress the formation of a 75–85-kDa PPAD species P. gingivalis sorting type II isolate 20663 was grown in the presence or absence of protease inhibitors as indicated. Growth medium fractions, containing OMVs, were analyzed by immunoblotting as indicated for . Molecular weights of marker proteins and different PPAD species are indicated.
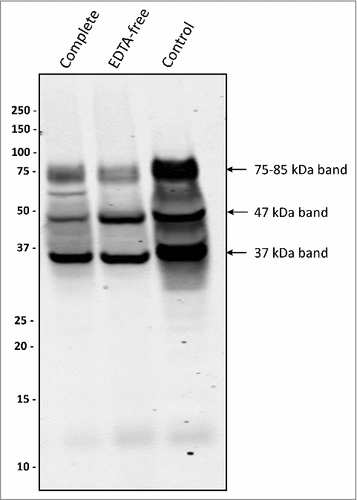
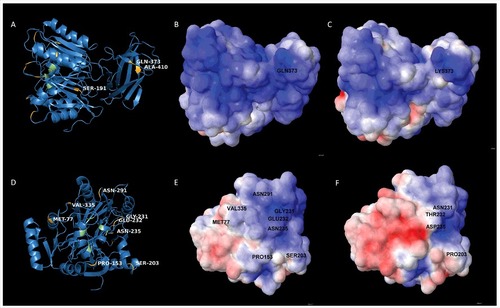
Figure 5. Position of amino acid substitutions in PPAD and their impact on the electrostatic potential of the protein (A) 3D-structural ribbon representation of the PPAD protein from P. gingivalis reference strain W83, showing in yellow surface-exposed amino acid residues that have been substituted in PPAD proteins from other P. gingivalis sorting type II isolates. Catalytic residues of PPAD are shown in green. (B and C) Electrostatic potential maps showing, respectively, the PPAD proteins of strain W83 and the sorting type II isolate MDS33 from the same perspective. The two maps display the difference in electrostatic potential (red represents -5 KT/e and blue +5 KT/e), and the respective Gln or Lys residues at position 373 are indicated. (D) 3D-structural ribbon representation of the PPAD protein from P. gingivalis strain W83, showing surface-exposed amino acid residues that have been substituted in other PPAD proteins (marked in yellow) of sorting type II isolates from the perspective of the catalytic site. The catalytic residues are marked in green. (E) Electrostatic potential map of the PPAD protein of strain W83, displaying all the residues subject to substitutions in sorting type II isolates. (F) Electrostatic potential map of the PPAD protein from the sorting type II isolate MDS33.