Figures & data
Table 1. The capsular serotype, MLST type and distribution of ten virulence factor genes in the 20 hypermucoviscous K. pneumoniae isolates with ultra-long viscous string (>20 mm).
Figure 1. Kaplan-Meier survival curves for K. pneumoniae RJF293 infected mice. Mice were infected with 103 CFU of different K. pneumoniae strains intraperitoneally. The previously reported hvKP strain NTUH-K2044 (ST23, K1 serotype), cKP strain HS11286 (ST11, KL103 serotype) and saline were applied as the controls. RJF293 showed virulence not statistically significant from that of NTUH-K2044 (p > 0.6, by log-rank test). No death of mice in the HS11286 or saline groups was observed during seven days.
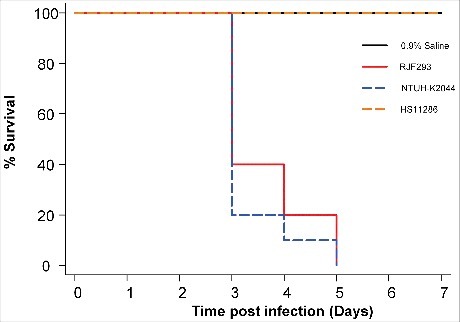
Figure 2. Putative virulence genes (gene clusters) detected among the 106 completely sequenced K. pneumoniae genomes (Supplementary Table S3). (A) A parsimony tree generated from 429,267 SNPs using kSNP3 for the 106 completely sequenced K. pneumoniae chromosomes and displayed by iTOL with midpoint rooting. The hvKP and cKP isolates listed in Supplementary Table S4 are highlighted by color (red or orange, hvKP; blue, cKP). (B) The virulence genes predicted in the RJF293 genome are listed as an example in Supplementary Table S5. The presence of genes in the chromosome and/or plasmid is indicated in different gray scales.

Table 2. Large genomic island-like regions (>10 kb) identified on the K. pneumoniae RJF293 chromosome.
