Figures & data
Figure 1. Cryptococcus neoformans yeast cells visualized by India Ink under phase contrast microscopy. India Ink particles are excluded from the dense PS capsule. A. Yeast cells cultured in an environment with optimal nutrition. B. Yeast cells cultured within mouse bone-marrow derived macrophages, experiencing starvation and oxidative stress from the phagolysosome for 24 h. Note the increased cell body, generation of what appears to be a large vacuole, and drastic increase in capsule size. Both images were obtained at 100x magnification with 2 × 2 binning.
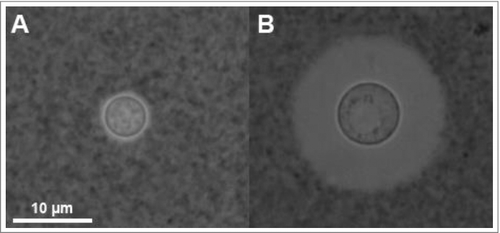
Figure 2. Knowledge of capsule structure as a function of dimension scale. Representative hyperbola of polysaccharide capsule structural understanding which is greatest at the smallest (molecular) and largest (cellular) levels. At the molecular level an in silico energy-minimized structure of the GXM M2 motif (see ) is estimated to cover a few nanometers in diameter. At the cellular level, capsule radii exist in the micrometer size range and are readily measured by negative staining and light microscopy (Scale bar in micrograph represents 10 µm). There is a knowledge gap in capsule structure at the macromolecular level or higher order structure (analogous to the secondary, tertiary and quaternary structural level of proteins) were many motifs are organized to form the overall capsular structure.
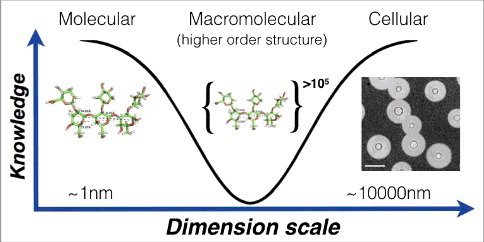
Figure 3. Illustration of the C. neoformans polysaccharide capsule structure. A. Cellular view of the polysaccharide capsule formed by a complex lattice-work of interconnected polymers that decreases in density, porosity and stiffness as it extends radially outward from the cell wall. B. Representative model of proposed polysaccharide lattice making up the capsular macromolecular structure involving interactions between GXM, GalXM, divalent cations, and mannoproteins. C. Structure of the six major motifs making up the GXM polysaccharide present in serotypes A, B, C, and D [Citation35]. These tetrasaccharide to octasaccharide motifs fit together to make up the megadalton GXM polymer. Each motif is color coded to match the lattice illustration in B. In C. neoformans serotype A motif 4 is not present [Citation35]. Motifs are represented using CFG nomenclature (mannose, green circles; xylose, orange 5-pointed start; glucuronic acid, blue/white diamond) along with chair pyranose rings and linkages.
![Figure 3. Illustration of the C. neoformans polysaccharide capsule structure. A. Cellular view of the polysaccharide capsule formed by a complex lattice-work of interconnected polymers that decreases in density, porosity and stiffness as it extends radially outward from the cell wall. B. Representative model of proposed polysaccharide lattice making up the capsular macromolecular structure involving interactions between GXM, GalXM, divalent cations, and mannoproteins. C. Structure of the six major motifs making up the GXM polysaccharide present in serotypes A, B, C, and D [Citation35]. These tetrasaccharide to octasaccharide motifs fit together to make up the megadalton GXM polymer. Each motif is color coded to match the lattice illustration in B. In C. neoformans serotype A motif 4 is not present [Citation35]. Motifs are represented using CFG nomenclature (mannose, green circles; xylose, orange 5-pointed start; glucuronic acid, blue/white diamond) along with chair pyranose rings and linkages.](/cms/asset/410ae2da-1c88-4911-8e87-52e92116e86d/kvir_a_1431087_f0003_oc.jpg)
