Figures & data
Figure 1. Lysozyme sensitivity of clinical ExPEC isolates and laboratory E. coli strains. (A) The densities of six E. coli strains in the late log phase of growth (OD600 = 2.0) were adjusted to 108 CFU/mL, and the cells incubated with different lysozyme concentrations (0–50 mg/mL) in a 96-well microtiter plate. The lysozyme sensitivity was determined based on MLC, which was the lowest concentration of lysozyme to lyse E. coli cells following a 24-h incubation at 37 °C. (B) The in vitro lysozyme killing assay. The degree of bacterial lysozyme sensitivity was calculated by dividing the CFU number prior to treatment by the CFU number after a 24-h exposure to lysozyme (N0/N). Data represent the mean ± standard deviation (SD) from three independent experiments. ##P < 0.01 by one-way ANOVA.
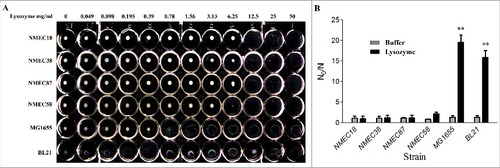
Table 1. E. coli strains and plasmids used in this study.
Table 2. Primers used in this study.
Table 3. Genetic loci disrupted by mini-Tn5 in the derivatives of strain NMEC38 Nalr.
Figure 2. Novel genes involved in the O-antigen biosynthesis contribute to ExPEC resistance to lysozyme. (A) Comparison of the O-antigen cluster in IHE3034 (NC_017628.1), MG1655 (NC_000913), and G1630 (GU299793) strains. Genes designated by purple are involved in sugar biosynthesis; green, genes involved in the O-antigen processing; blue, genes encoding glycosyltransferase enzymes; and black outline, genes that have been found to be associated with lysozyme resistance in the current study. The linear representation of the genetic comparison was generated using Easyfig version 2.1. (B) A silver-stained polyacrylamide gel after SDS-PAGE (top) and generalized LPS structures (bottom). (C) Western blotting profiles of LPS probed with the anti-O18 serum. Compared with the wild-type strain, the banding pattern in the O-antigen region was altered in all mutant strains. The complemented strains had similar banding patterns to the wild type, except for the complemented strain ΔECOK1_2265. Rha: rhamnose; Gal: galactose; Glc: glucose; Kdo: 3-deoxy-d-manno-oct-2-ulosonic acid; Hep: l-glycero-d-manno-heptose; GlcNAc: N-acetylglucosamine.
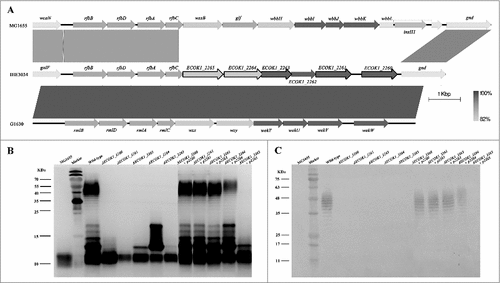
Figure 3. The in vitro lysozyme killing assay and membrane integrity tests. (A) Lysozyme sensitivity of the wild-type, mutant, and complemented strains was determined by an in vitro killing assay. The strain compromised viability levels (N0/N) under the indicated conditions were analyzed as in . (B) Membrane integrity of the wild-type, mutant, and complemented strains determined by PI staining. (C) Membrane integrity determined by a protein leakage assay. Data represent the mean ± standard deviation (SD) from three independent experiments. #P < 0.05, ##P < 0.01 by one-way ANOVA.
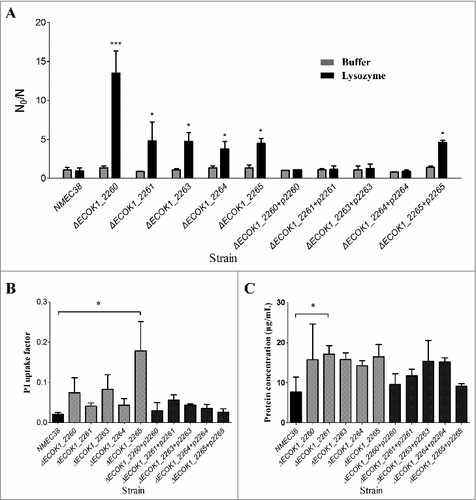
Figure 4. The in vitro inhibition of lysozyme activity by LPS and the O-specific polysaccharide from NMEC38 wild type. (A) In vitro inhibition of the lysozyme killing activity. To determine the effect of LPSNMEC38 on lysozyme killing activity, ΔECOK1_2265 strain was incubated with lysozyme in the presence or absence of diluted LPSNMEC38 for 24 h. The control cells were incubated in 1 mM Tris-HCl (pH 7.2). Inactivation levels (N0/N) were determined after the incubation period. (B) LPS without the O-specific polysaccharide and O-specific polysaccharide alone cannot inhibit the bactericidal activity of lysozyme. Compromised viability (N0/N) were determined in the absence and presence of 2.0 mg/mL of LPS or O-SP. (C) In vitro inhibition of lysozyme hydrolysis activity by different concentrations of LPS. M. lysodeikticus cell lysis was determined as the decrease in optical density at 600 nm (OD600) over time, in the presence of 4 μg/mL of lysozyme, and in the absence or presence of LPS from the wild type, NMEC38. (D) The ability of LPS lacking the O-specific polysaccharide to inhibit the hydrolytic activity of lysozyme is substantially reduced. M. lysodeikticus cells were suspended in the presence of 4 μg/mL of lysozyme in the absence or presence of 100 μg/mL of LPS from wild-type, mutant, and MG1655 strains, and the experiment was performed as in (C). Data represent the mean ± standard deviation (SD) from three independent experiments. #P < 0.05; ##P < 0.01; ###P < 0.001 by one-way ANOVA. LSM, Lysozyme; O-SP, O-specific polysaccharide.
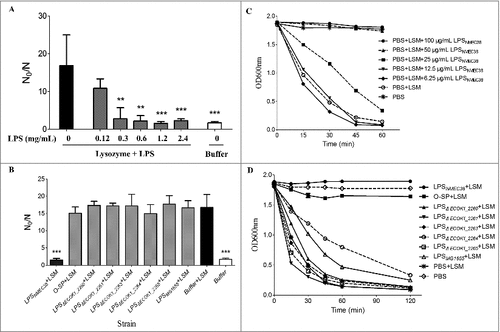
Figure 5. Analysis of LPSNMEC38 and lysozyme by gel filtration chromatography. (A) In the absence of LPS, lysozyme was eluted in fractions no. 13–19. SDS-PAGE and western blotting were used to detect the eluted lysozyme. (B) In the absence of lysozyme, LPS was eluted in two peaks, as determined by photometry, but only the first peak contained LPS, as indicated by SDS-PAGE (LPS silver-staining) (fractions 3–5). (C) Co-incubation of lysozyme with LPS resulted in the presence of lysozyme in the forward-shifted peak that contained fractions A4–A7. i) Gel filtration chromatography analysis of the lysozyme-LPS complex. ii) Presence of lysozyme in the peak containing fractions A13–A19, as demonstrated by SDS-PAGE (LPS silver-staining) and western blotting (with anti-lysozyme antibody). iii) Presence of lysozyme in the pronouncedly forward-shifted peak containing fractions A4–A7 (lysozyme presence in the peaks containing fractions 13–19 in ) shown by SDS-PAGE (LPS silver-staining) and western blotting (with anti-lysozyme antibody).
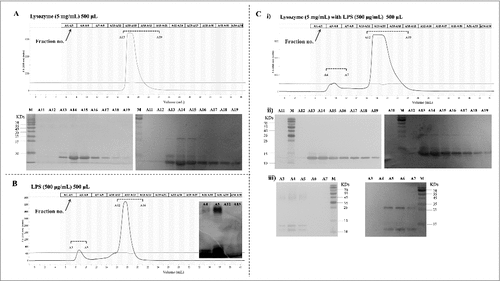
Figure 6. The O-specific polysaccharide of ExPEC can directly bind to lysozyme. (A) Gel filtration chromatogram of lysozyme. (B) Gel filtration chromatogram of the O-specific polysaccharide. (C) Co-incubation of lysozyme with the O-specific polysaccharide resulted in only one peak in the chromatogram; the peak was pronouncedly forward-shifted in comparison with the peak of the O-polysaccharide in (B). O-SP, O-specific polysaccharide.
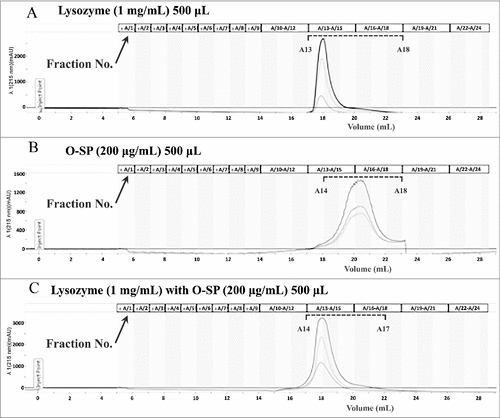
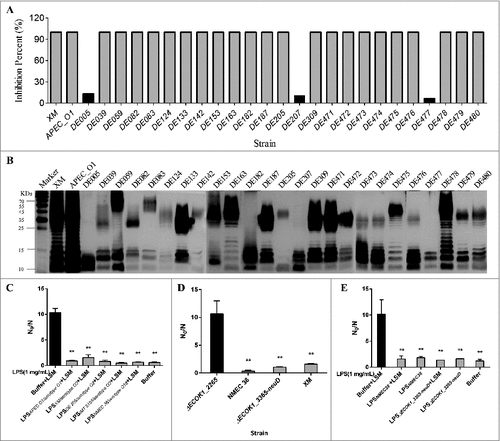
Figure 7. (A) Percent inhibition of the hydrolytic activity of lysozyme. Strains that harbor intact LPS (24) completely inhibited the hydrolytic activity of lysozyme, while the inhibition was almost unnoticeable with strains lacking the O-specific polysaccharide; (B) LPS profiles of 27 ExPEC strains analyzed by SDS-PAGE and silver-staining; (C) the purified LPS from ExPEC with O1, O2, and O18 could protect mutant strain ECOK1_2265 from the bactericidal activity of lysozyme; (D) deletion of capsule biosynthesis genes (ΔECOK1_3365-neuD) from wild type strain did not significantly reduce the ExPEC's resistance to lysozyme, while deletion of O antigen biosynthesis gene (ΔECOK1_2265) significantly reduced ExPEC's resistance to lysozyme, XM: Extraintestinal pathogenic E. coli wild type strain ExPEC XM with serotype of O2 (control); and (E) purified LPS from mutant strain with capsule biosynthesis gene (ΔECOK1_3365-neuD) deletion showed that capsule polysaccharides were not necessary for the LPS-mediated protection of ExPEC from the bactericidal activity of lysozyme. Data represent the mean ± standard deviation (SD) from three independent experiments. #P < 0.05; ##P < 0.01; by one-way ANOVA. LSM, Lysozyme.