Figures & data
Figure 1. Virulence of M. circinelloides CBS277.49 and NRRL3631 strains in neutropenic murine host. Survival of mice infected (A) intravenously (i.v.) with 1 × 106 and 1 × 105 CFU of fungal strains and (B) intranasally (i.n.) with 5 × 107 CFU of fungal strains show significant lower virulence of NRRL 3631 (#) (p < 0.05). (C) Fungal biomass quantification of liver, brain, kidney lung and spleen 3 days after i.v. infection with 1 × 106 CFU/animal of CBS277.49 and NRRL3631 using real-time PCR. Data represent µg of fungal biomass respect 1 g of mice tissue biomass. Significant differences are indicated (#). Bars indicate the standard deviation from independent experiments.
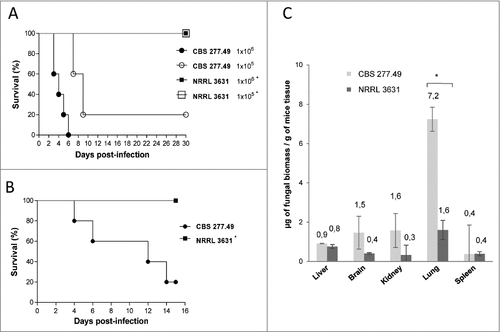
Figure 2. Fungal strains phenotypes. Small and large spores of CBS277.49 strain exhibit differences in (A) germination kinetics and (B) similar virulence capacity in murine model infection. (C) Fungal colonies from CBS277.49 and NRRL3631 M. circinelloides strains grown for 2–3 days at 30°C on SM plates containing sodium dodecyl sulphate (SDS) or calcofluor white (CFW) and 5 days at 35ºC and 37ºC, inoculating 103, 102 and 10 fresh spores. (D) Percentage of chitin and chitosan respect to initial mycelial dry weight.
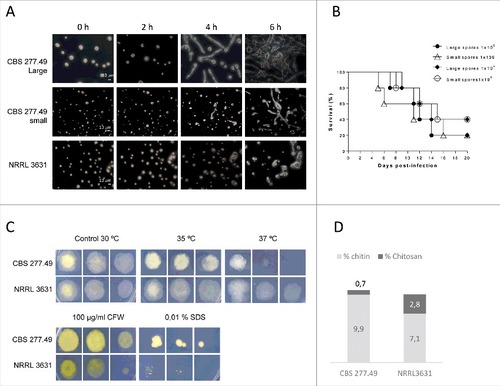
Table 1. Assembly and mapping statistics of M. circinelloides NRRL3631 genome sequencing.
Figure 3. Genes identified in the genome comparison study clustered in functional categories based on KOG classification. (A) Absent genes and (B) discontiguous genes in NRRL3631 strain.
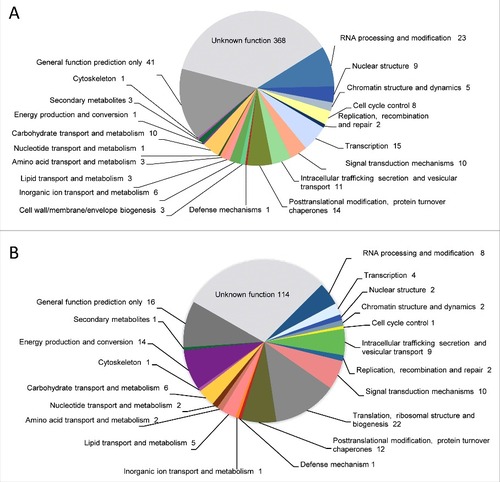
Table 2. Eukaryotic Orthologous Groups (KOG) classification of absent or discontiguos genes detected in NRRL3631 genome in comparison to CBS277.49. A total of 395 sequences assigned to KOG classifications within the principal 17 categories are shown.
Table 3. Genes coding for extracellular enzymes identified in the genome comparison study among CBS277.49 and NRRL3631 genomes as absent, or discontiguous sequences in NRRL3631. aNumber of duplicated genes in CBS277.49 M. circinelloides genome based upon BLAST with E-value < E−50 and ≥ 70% identity. bDistribution of orthologues within the fungi species based upon BLAST with E-value < E−22 and ≥ 50% identity. Ac: Absidia glauca, Cc: Choanephora cucurbitarum, Cg: Colletotrichum gloeosporioides, Lc: Lichtheimia corymbifera Lr: Lichtheimia ramosa, Ma: Mucor ambiguos, Mc: Mucor circinelloides, Me: Morteriella elongata, Pb: Phycomyces blaskespeanus, Pp: Parasitella parasítica, Rm: Rhizopus microsporus, Ri: Rhizopus irregularis, Rd: Rhizopus delemar, Ro; Rhizopus oryzae, Um: Ustilago maydis.
Figure 4. Disruption of 112092 and 108920 genes. (A) Targeted replacement strategy using a vector generated by fusion PCR with the pyrG gene as selective marker. Black arrowheads indicate the primer pairs used for amplification of DNA fragments. Probes are indicated (dashed bar). (B) Southern analysis of gDNAs from M. circinelloides CBS277.49 and transformants. DNAs were digested with EcoR V and Sma I to detect 112092 and 108920 gene deletions, respectively.
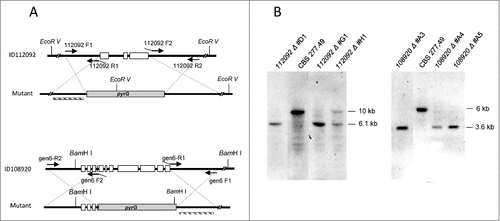
Figure 5. Virulence of knockout mutants in neutropenic mice infection. (A) and (B) Survival of mice infected intravenously (i.v.) with 1 × 105 CFU and (C) mice infected intranasally with 5 × 107 CFU of fungal strains. Data showed attenuated virulence of mutants in ID112092 and ID108920 genes. Significant differences are indicated (#).
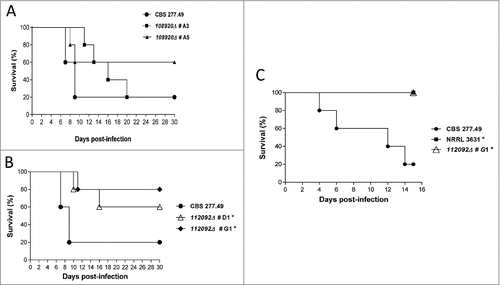
Table 4. Oligonucleotides used in this study. Italics and lower case indicate nucleotide sequences added for fusion purposes. Positions are referred to the start codon, (+) downstream or (-) upstream of ATG. Orientation is indicated, (s) sense, (as) antisense.
