Figures & data
Figure 1. Quantitative RT-PCR demonstrated that expression of the ORF19.1539, ORF19.1725, ORF19.2430, ORF19.2691 and ORF19.5557 genes in white cell pheromone response were regulated by Cph1 in P37005. Expression was analyzed and compared between the wild-type and cph1Δ with or without α-pheromone peptide treatment. Expression was normalized to that of the ACT1 gene. Values are the mean ± SD of three experimental replicates, and two technical repeats were performed for each experimental replicate. ##: P < 0.01.
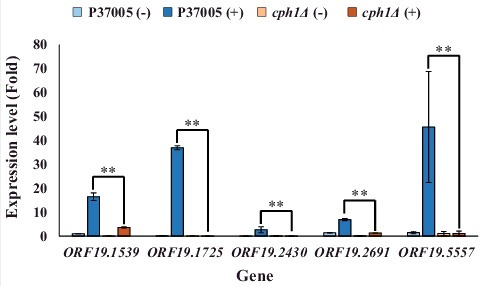
Figure 2. ORF19.1539, ORF19.1725, ORF19.2430 or ORF19.5557 in C. albicans P37005 are specifically involved in white cell pheromone response but are dispensable in opaque cell pheromone response. (A) White cells of P37005 cultured in Lee's medium in the presence or absence of pheromone (MFα) in plastic dishes. Images were taken after PBS washing (top panel), and the remaining adhered cells were quantitated (bottom panel). (B) Opaque cells of the wild-type and mutant strains (MTLa/a; SAT−, Arg+) were crossed with the MTLα/α DSY211 strain (SAT+, Arg−) on Spider medium for 48 hr. Cells were plated onto selective media (SAT+, Arg−) to quantitate mating efficiencies. The cph1Δ strain of P37005 served as a negative control. Values are the mean ± SD of three experimental replicates, and two technical repeats were performed for each experimental replicate. #: P < 0.05; ##: P < 0.01.
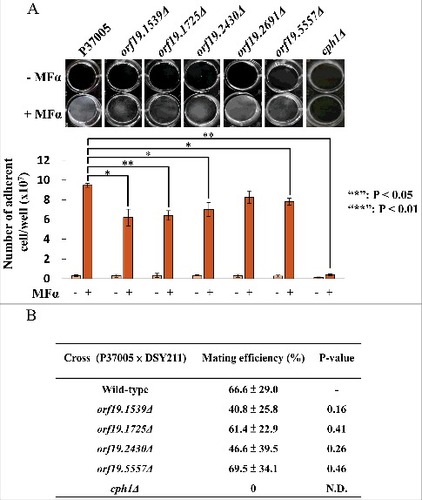
Figure 3. ORF19.1725 is also regulated by Tec1 and is required for the formation of conventional biofilms. (A) Analysis of five candidate genes in a conventional biofilm assay on silicone squares revealed that the presence of the ORF19.1725 gene is necessary for the development of conventional biofilms. The graph shows the mean ± SD of three experimental replicates, and two technical repeats were performed for each experimental replicate. “##”: P < 0.01. (B) Quantitative RT-PCR revealed that ORF19.1725 expression was regulated by Tec1. Expression was analyzed and compared between the wild-type and tec1Δ in response to pheromone treatment. Expression was normalized to that of the ACT1 gene. Values are the mean ± SD of three experimental replicates, and each replicate represents two technical repeats. “##” represents P < 0.01. – and + indicate with or without pheromone treatment, respectively.
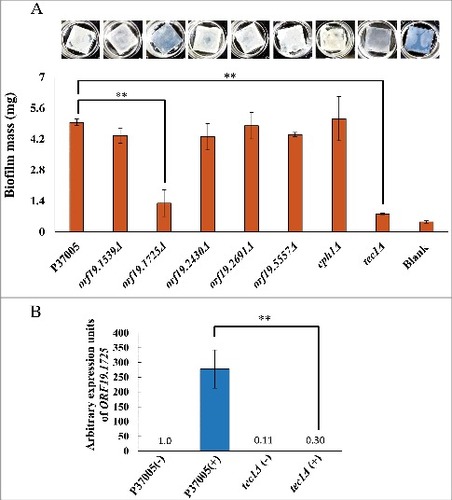
Figure 4. The orf19.1725Δ strain was defective in hyphal formation. (A) Cells of the wild-type, orf19.1725Δ and complemented strains were grown in YPD medium with or without 50% bovine serum. The representative images show that ORF19.1725 gene deletion caused a severe defect in hyphal development. Four fields were checked, and at least 100 cells were counted in each field of every C. albicans strain. Ratios of hyphae formation are displayed below each image. “##” represents P < 0.01 and “###” represents P < 0.001. (B) The wild-type and the complemented strains clearly exhibited clumps after 12 hr of culturing in YPD liquid media at 37°C, whereas the orf19.1725Δ strains did not. Hyphae were observed and examined under a light microscope after the clumps had been collected by centrifugation. Left panel: without serum treatment; right panel: YPD supplemented with 50% bovine serum. Scale bar: 50 μm. (C) Growth curves of C. albicans strains at 37°C. Overnight cultures of YPD-grown C. albicans cells were diluted to an OD600 of 0.1 in fresh YPD liquid media. Growth rates were monitored every 4 hr using a Biowave density meter. Values are the mean ± SD of three experimental replicates, and two technical repeats were performed for each experimental replicate. “##” represents P < 0.01 and “###” represents P < 0.001.
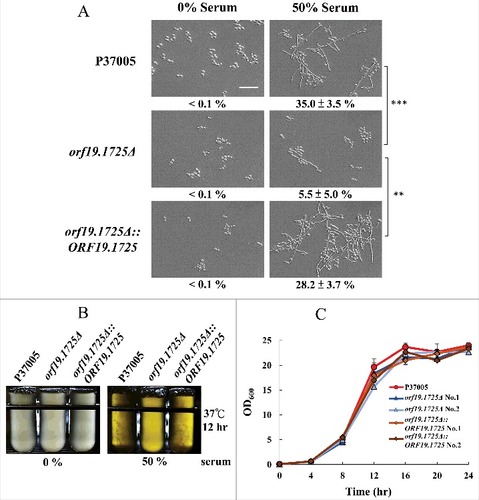
Figure 5. The C. albicans Orf19.1725 protein potentially functions as an adhesin. (A) Analysis of Orf19.1725 reveals three conserved features, a putative signal peptide, four different tandem repeats (TR1, TR2, TR3 and TR4) and a hypothetical ω site. (B) Protein sequence of C. albicans orf19.1725. The arrow between the 26th (Gly) and 27th (Leu) amino acids indicates the likely cleavage site for the signal peptide (http://www.cbs.dtu.dk/services/SignalP/). The other, at the position of the 700th amino acid (Asp), was predicted to be an ω site with lower specificity (http://gpcr.biocomp.unibo.it/predgpi/). (C) Sequence motifs of four different tandem repeats of Orf19.1725 (http://weblogo.threeplusone.com/create.cgi).
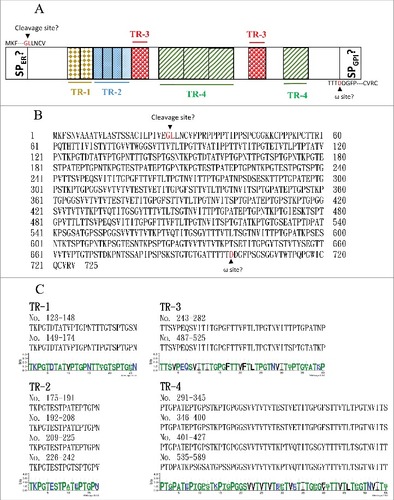
Figure 6. ORF19.1725 is involved in cell adhesion to zebrafish embryos. (A) Embryos were co-incubated with 5 × 105 C. albicans cells of P37005 (a, g, m), orf19.1725Δ (b, c, h, i, n, o) and orf19.1725Δ::ORF19.1725 (d, e, j, k, p, q) for 4 hr. The infected embryos were then washed with egg water to remove non-adhered cells. Images were taken by a Leica TCS SP5 II inverted microscope. The images in the middle panel (g, h, i, j, k, l) are focused on the outside of the embryo surface. The images in the bottom panel (m, n, o, p, q, r) are focused on the inner side of the embryo surface. Embryos without infection by C. albicans cells served as negative controls (f, l, r). White arrows indicate hyphae on or in the embryo surfaces. (B) Cell quantitation revealed a significantly lower number of adhesive cells of orf19.1725Δ on zebrafish embryo surfaces than of the wild-type or the complemented strains. Infected embryos were washed twice with 40 ml of egg water to remove non-adhered cells. The embryo eggs were then disrupted by a FastPrep-24 instrument (MP Biomedicals, Illkirch, France) and plated on YPD plates to quantitate the number of adhered cells. Values are the mean ± SD of three experimental replicates, and two technical repeats were performed for each experimental replicate “##” represents P < 0.01.
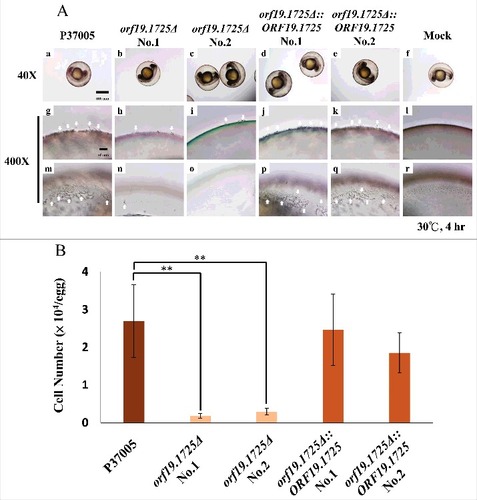
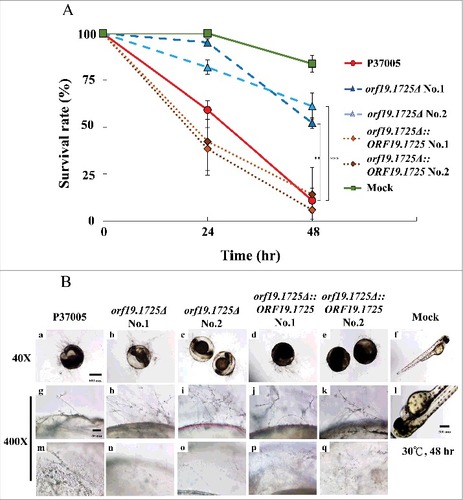
Figure 7. ORF19.1725 deletion strains had reduced virulence. (A) Survival curves of zebrafish embryos during infection with C. albicans cells revealed that Orf19.1725 is involved in pathogenicity. Twenty embryos in each plastic well were co-incubated with 5 × 105 C. albicans cells. Infected embryos were washed and transferred to sterilized egg water. Survival rate was determined after calculating the number of infected zebrafish embryos that retained a heartbeat divided by the total embryo number. Values are the mean ± SD of three experimental replicates, and each replicate represents two technical repeats. “##” represents P < 0.01 and “###” represents P < 0.001. (B) Representative images of embryos infected by C. albicans after 48 hr revealed that orf19.1725Δ exhibited slower infection progress, less penetration ability and a clearer embryo background (b, c, h, i, n, o) than the wild-type strain (a, g, m) and the complemented strains (d, e, j, k, p, q) did. Embryos without infection by C. albicans cells served as mock controls (f, l) and hatched into newborn zebrafish. The images in the middle panel (g, h, i, j, k, l) are focused on the outside of the embryo surface. The images in the bottom panel (m, n, o, p, q, r) are focused on the inner side of the embryo surface.