Figures & data
Figure 1. Brassica plant root affected by Plasmodiophora brassicae. A. Canola root with typical galls after 1 month of inoculation with P. brassicae resting spores. B. Life cycle of P. brassicae showing the steps involved in infection through to the production of secondary plasmodia in the host plant cortical cells. Scheme based on that of Kageyama and Asano [7], representing spindle-shaped resting spores, biflagellate primary zoospores, zoospores, and primary and secondary plasmodia (oval black figure in root hairs and cortical cells, respectively). Further steps in P. brassicae´s life cycle, such as the formation of resting spores in cortical cells and its ejection to the soil, are not shown in this scheme.
![Figure 1. Brassica plant root affected by Plasmodiophora brassicae. A. Canola root with typical galls after 1 month of inoculation with P. brassicae resting spores. B. Life cycle of P. brassicae showing the steps involved in infection through to the production of secondary plasmodia in the host plant cortical cells. Scheme based on that of Kageyama and Asano [7], representing spindle-shaped resting spores, biflagellate primary zoospores, zoospores, and primary and secondary plasmodia (oval black figure in root hairs and cortical cells, respectively). Further steps in P. brassicae´s life cycle, such as the formation of resting spores in cortical cells and its ejection to the soil, are not shown in this scheme.](/cms/asset/01c07563-ed09-4d29-a051-ad5311bdeb4c/kvir_a_1504560_f0001_oc.jpg)
Table 1. Steps to identify putative effectors within the secretome of P. brassicae in the European strain Pbe3 and the Canadian strain Pb3.
Table 2. Bioinformatics tools used in a coherent pipeline to identify putative effectors of P. brassicae.
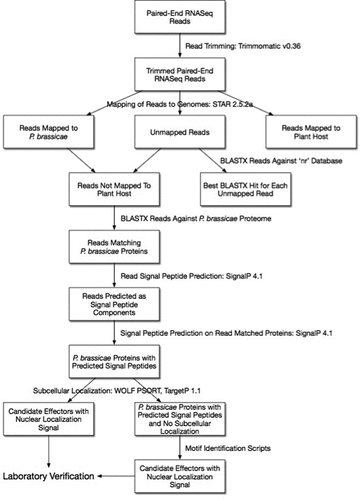
Figure 2. Coherent pipeline to identify putative effector proteins of Plasmodiophora brassicae. The pipeline assumes: (1) Researchers are starting with RNA-Seq reads from a host plant infected with P. brassicae; (2) The draft genomes available for P. brassicae, other plasmodiophorids, oomycetes, and other plant pathogens are used; (3) Motifs mentioned in the review or structural similarities with previously described effector proteins were identified.