Figures & data
Table 1. Genomic features and properties of the newly sequenced genome of B. melitensis strain Rev.1, as compared with the known genome sequence of B. melitensis strain 16M.
Figure 1. Whole-genome alignment of the B. melitensis 16M and Rev.1 strains. Mauve alignment of both chr I (A) and chr II (B) from the complete genomes of B. melitensis rev.1 (bottom panel) and 16M (top panel) are shown. The colored blocks indicate individual locally collinear blocks (LCBs). Same-colored blocks indicate homologous regions. The homologous LCBs are connected among the two strains. Same-colored blocks on opposite sides of the line indicate inversions. The boundaries of colored blocks indicate the breakpoints of genome rearrangement. (C) A dot plot comparing chr II of the Rev.1 and 16M strains using the LAST local alignment software (default parameters) [Citation60]. The inversion found in the Rev.1 genome is indicated in red. (D) A scaled diagram of the ORFs located upstream and downstream of the inversion site on chrII of B. melitensis.
![Figure 1. Whole-genome alignment of the B. melitensis 16M and Rev.1 strains. Mauve alignment of both chr I (A) and chr II (B) from the complete genomes of B. melitensis rev.1 (bottom panel) and 16M (top panel) are shown. The colored blocks indicate individual locally collinear blocks (LCBs). Same-colored blocks indicate homologous regions. The homologous LCBs are connected among the two strains. Same-colored blocks on opposite sides of the line indicate inversions. The boundaries of colored blocks indicate the breakpoints of genome rearrangement. (C) A dot plot comparing chr II of the Rev.1 and 16M strains using the LAST local alignment software (default parameters) [Citation60]. The inversion found in the Rev.1 genome is indicated in red. (D) A scaled diagram of the ORFs located upstream and downstream of the inversion site on chrII of B. melitensis.](/cms/asset/9606057e-9a32-4db1-8698-eafbc3554984/kvir_a_1511677_f0001_oc.jpg)
Table 2. The RIs and RDs within the rev.1 genome.
Figure 2. Regions of insertions within the MerR (A) and BMEI1570 (B) proteins of the B. melitensis Rev.1 strain. Pairwise alignment between the Rev.1 (top) and 16M (bottom) sequences was visualized using the Clustal X color scheme for residues coloring [Citation63]. (C) Crystal structure of the E. Coli copper efflux regulator homodimer (4WLS). DNA is shown as white surface. The inserted sequence 1 (RI1) is shown in red. (D) Crystal structure of the T. maritima glycerate kinase (2B8N). The inserted sequence 2 (RI2) is shown in yellow. Active site residues are shown as spheres.
![Figure 2. Regions of insertions within the MerR (A) and BMEI1570 (B) proteins of the B. melitensis Rev.1 strain. Pairwise alignment between the Rev.1 (top) and 16M (bottom) sequences was visualized using the Clustal X color scheme for residues coloring [Citation63]. (C) Crystal structure of the E. Coli copper efflux regulator homodimer (4WLS). DNA is shown as white surface. The inserted sequence 1 (RI1) is shown in red. (D) Crystal structure of the T. maritima glycerate kinase (2B8N). The inserted sequence 2 (RI2) is shown in yellow. Active site residues are shown as spheres.](/cms/asset/29da4b4a-d43b-4428-82e9-3645a4efd2cd/kvir_a_1511677_f0002_oc.jpg)
Table 3. List of ORFs that contain SNPs and encode reported under-expressed proteins in the B. melitensis strain Rev.1, as compared with strain 16M.
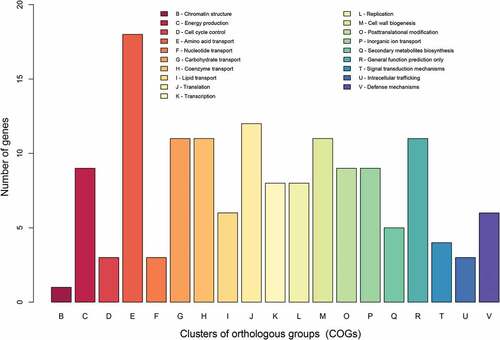
Figure 4. COG-based functional categories of the 160 ORFs that are different between the vaccine Rev.1 and the virulent 16M B. melitensis strains.