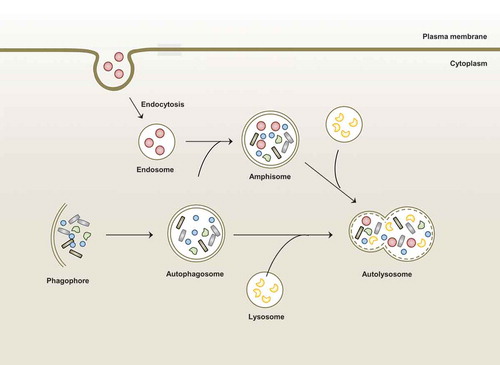
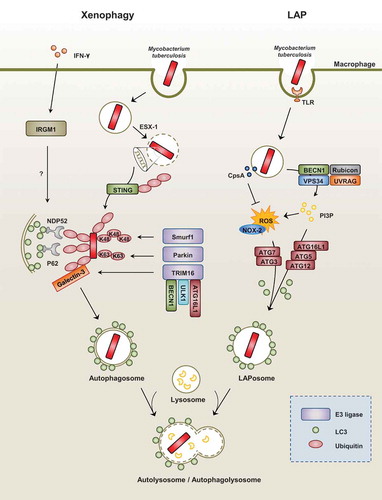
Figures & data
Table 1. Potential adjunctive anti-TB therapeutics targeting autophagy.
Table 2. Clinical studies of autophagy-adjunctive therapeutics for tuberculosis.