Figures & data
Figure 1. TGEV infection activated the NF-κB pathway.
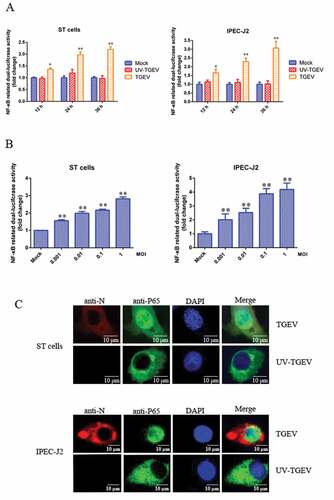
Figure 2. TGEV infection induced inflammation through the activation of the NF-κB pathway.
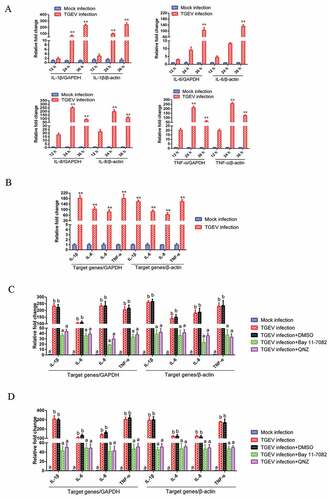
Figure 3. The effects of the NF-κB signaling pathway on TGEV replication.
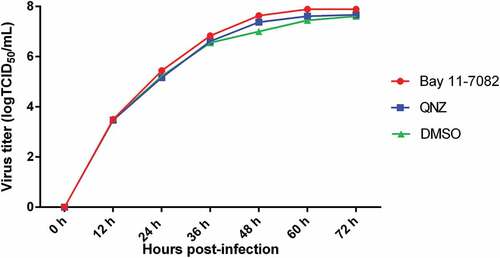
Figure 4. TGEV Nsp2 activated NF-κB.
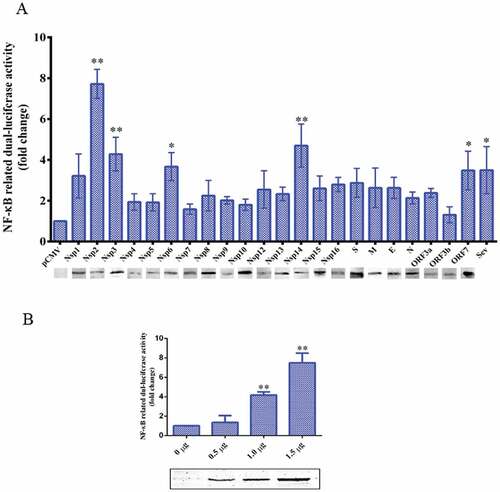
Figure 5. TGEV Nsp2 induced the degradation of IκBα and the nuclear translocation of p65.
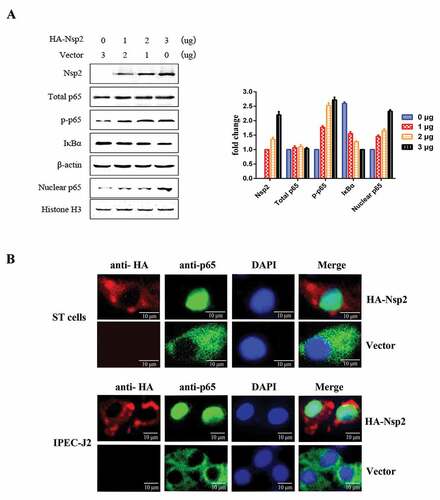
Figure 6. Nsp2 expression enhanced the expression of NF-κB-regulated pro-inflammatory genes.
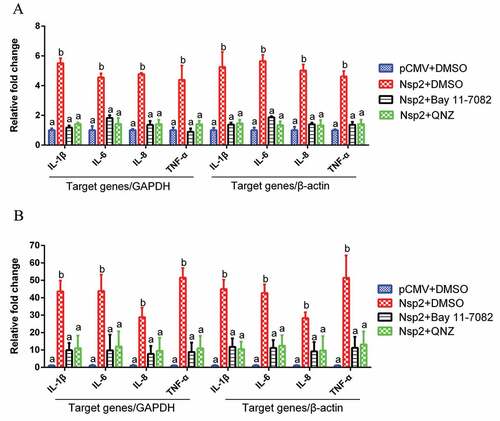
Figure 7. The amino acids 1–120 of Nsp2 were responsible for the activation of the NF-κB signaling pathway.
Table 1. Primers used for vector construction.
Table 2. Primers used for RT-PCR.
