Figures & data
Figure 1. Schematic illustration of the molecular mechanism of autophagy. Autophagosome formation is initiated by the nutrient-sensing kinases (i.e., mTORC1 and AMPK) and through the function of three protein complexes (i.e., ULK1/2 complex, class III PI3K complex, and ATG2-WIPI complex) and two ubiquitin-like conjugation systems (i.e., ATG12-ATG5-ATG16L1 and LC3-PE). Once formed, the autophagosome then fuses with the lysosome to produce an autolysosome. Autophagosome can also merge with the endosome to form an amphisome, which then fuse with a lysosome for cargo degradation. The fusion between an autophagosome and a lysosome is regulated by multiple proteins, particularly the (STX17-SNAP29-VAMP8) SNARE complex. Selective cargo degradation is mediated by autophagy receptors, such as SQSTM1, NBR1, and CALCOCO2, which harbor a UBA domain for substrate recognition, and a LIR for binding to autophagosome-anchored LC3-II, thereby bridging the substrate to the autophagosome for degradation. mTORC1, mechanistic target of rapamycin complex 1; AMPK, AMP activated protein kinase; ATG, autophagy-related; ULK, uncoordinated (UNC)-51-like kinase; RB1CC1, RB1 inducible coiled-coil 1; PI3KC3, phosphatidylinositol 3-kinase catalytic subunit type 3; PIK3R4, phosphoinositide 3-kinase regulatory subunit 4; AMBRA1, activating molecule in Beclin-1-regulated autophagy protein 1; PI3P, PtdIns(3)P; WIPI, WD-repeat protein interacting with phosphoinositides; LC3, microtubule-associated protein light chain 3; PE, phosphatidylethanolamine; SQSTM1, sequestosome 1, NBR1, neighbor of BRCA1; CALCOCO2, calcium binding and coiled-coil domain-containing protein 2; SNAP29, synaptosomal-associated protein 29; STX17, syntaxin 17; VAMP8, vesicle-associated membrane protein 8; LIR, LC3-interacting region; UBA, ubiquitin-associated; Ub, ubiquitin.
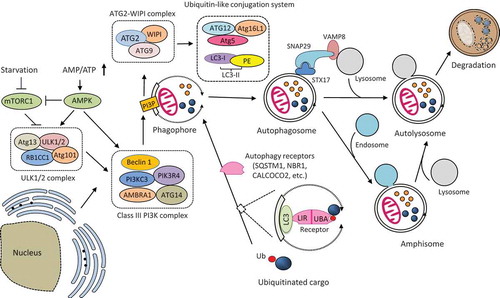
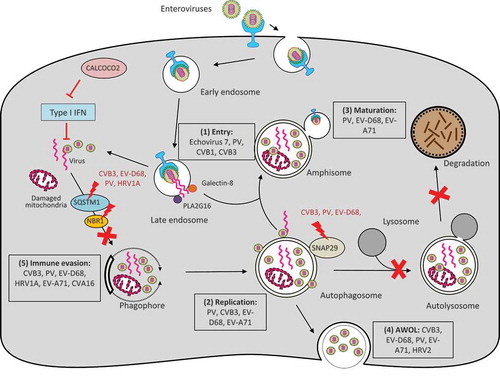
Figure 2. Interplay between enteroviruses and the host autophagy pathway. Major members of enteroviruses have been shown to subvert the host autophagy machinery at different phases of viral life cycle to enhance viral growth and for immune escape. (1) Viral entry. Autophagy core proteins benefit echovirus 7 entry, likely through regulation of the endocytic pathway. During PV, CVB1, and CVB3 infection, PLA2G16 is recruited to the fragmented endosome to antagonize the function of galectin-8 in initiating autophagic degradation of viral RNA genome. (2) Viral replication. Autophagosome-lysosome fusion is blocked during CVB3, PV, and EV-D68 infection through viral protease 3C-induced cleavage of SNAP29. Accumulation of autophagosomes appears to favor viral replication by offering replication membranes for PV, CVB3, EV-D68, and EV-A71. (3) Viral maturation. Formation of acidified amphisomes is required for EV (e.g., PV, EV-D68, EV-A71) maturation to generate infectious viruses. (4) Viral release through AWOL. EVs (e.g., PV, EV-D68, EV-A71, CVB3, and HRV2) utilize autophagosomes as envelopes for non-lytic viral exit. (4) Immune evasion. Autophagy receptor SQSTM1 is cleaved by viral protease 2A during CVB3, PV, EV-D68, and HRV1A infection (cleavage of NBR1 is also observed during CVB3 infection) to counteract virophagy-mediated clearance of viral particles/components. CALCOCO2 exhibits a pro-viral function toward CVB3 via inhibiting type I IFN signaling. Enhanced autophagy during EV-A71 and CVA16 infection was shown to benefit viruses by suppressing TLR7-mediated type I IFN signaling. PV, poliovirus; CVB, coxsackievirus B; CVA, coxsackievirus A; EV, enterovirus; HRV, human rhinovirus; SNAP29, synaptosomal-associated protein 29; SQSTM1, sequestosome 1, NBR1, neighbor of BRCA1; CALCOCO2, calcium binding and coiled-coil domain-containing protein 2; AWOL, autophagosome-mediated exit without lysis; IFN, interferon.