Figures & data
Figure 1. Several signaling pathways involved in appressorium formation and pathogenicity in Magnaporthe oryzae.
a. The Pmk1 MAPK and autophagy pathways are involved in different stages of the plant infection process.
b. Summary of the detection and transmission of fungal pathogenesis-related proteins to the outside signals. Pth11 can recognize surface signals, such as surface hydrophobicity and hardness, and activate downstream Mst11-Mst7-Pmk1 MAPK and endosomal pathways. These two pathways regulate appressorium formation, penetration, and invasive growth. TOR (Target of rapamycin) recognizes extracellular signals, such as nitrogen, glucose and some environmental stresses, and regulates autophagy by controlling the Atg1-Atg13-Atg17 complex.
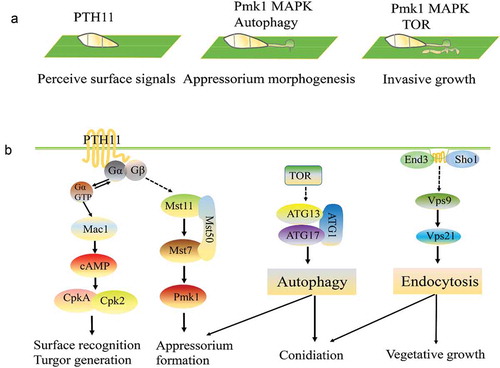
Table 1. Core autophagy genes related to pathogenicity in filamentous fungi.
Table 2. New factors that regulate autophagy in filamentous fungi.
Figure 2. Autophagy process in filamentous fungi.
After the induction of autophagy, an initial sequestering phagophore is assembled at the phagophore assembly site (PAS). Expansion and curvature of the phagophore leads to engulfment of the cargo (cytoplasm containing proteins and organelles) into the double-membraned autophagosome. The fusion of the autophagosomal outer membrane with the vacuolar membrane results in the release of the autophagic body, which is surrounded by the inner autophagosomal membrane. Autophagic bodies and their cargoes are degraded by hydrolytic enzymes, and the degraded products are exported into the cytoplasm for reuse.
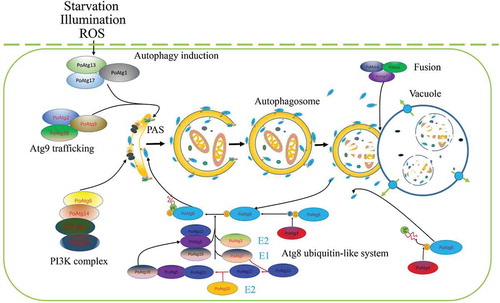
Figure 3. Autophagy mediates the infection of filamentous fungi and cell survival.
a. The deletion of any of the core ATG genes in filamentous fungi results in defects in fungal penetration into the host. The autophagy-deficiency mutants lose their ability to cause disease in their host plants.
b. In plate culture, the cells of autophagy-deficiency mutants lose their ability to recycle the materials produced by the cells. As a result, nutrients can no longer be transmitted between hyphal cells, and the middle sections of old hyphae collapse due to nutrient depletion.
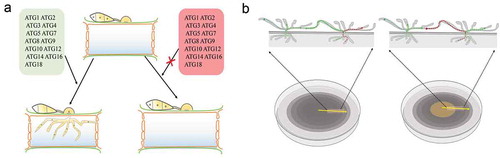
Table 3. Proteins with dual roles in autophagy and endocytosis.
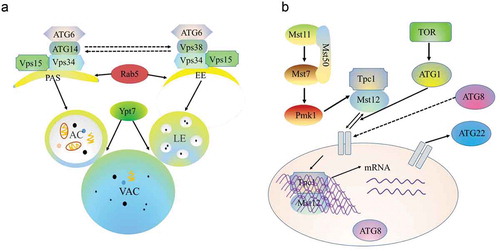
Figure 4. Crosstalk among autophagy and endocytosis and the Pmk1 MAPK pathway.
a. Vps34 recruits Atg6 under the action of Vps38 to induce endosomal endocytosis. Simultaneously, Atg6 is recruited by Vps34 under the action of Atg14 to target the PAS, which participates in the autophagy pathway. Both autophagosome and endosome fusion with the vacuole require the Ypt7 protein. PAS, phagophore assembly site; EE, early endosome; AC, autoplastic vacuole; LE, late endosome; VAC, vacuole.
b. In M. oryzae, environmental signals trigger the Pmk1 kinase cascade to control autophagy and activate Tpc1 function. Nuclear-localized Tpc1 then activates the transcription of genes required for polar growth, autophagy, and glycogen degradation.