Figures & data
Figure 1. (a) Neighbor joining tree of the IDV HEF gene reconstructed using a p-distance model implemented in MEGA7. (b) PCA of IDV according to different clades and hosts. The D/660, D/Japan and D/OK clades and swine and bovine hosts are represented in light blue, yellow, dark blue, grey, and orange, respectively.
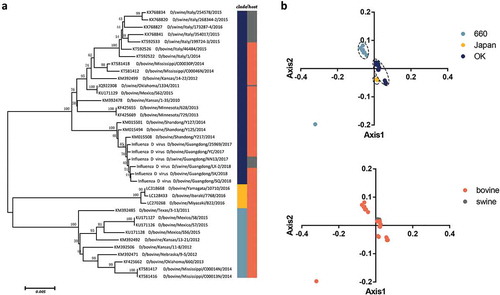
Table 1. The RSCU value of 59 codons encoding 18 amino acids according to clades and hosts of HEF gene. The optimal codons are shown in bold.
Figure 2. ENC values of the HEF gene of different clades and hosts. The D/660, D/Japan, and D/OK clades and swine and bovine hosts are represented in light blue, yellow, dark blue, grey, and orange, respectively.
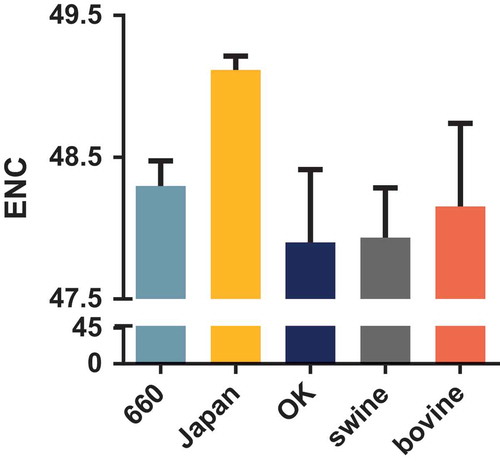
Figure 3. (a, b). ENC-plot analysis of the HEF gene, with ENC against GC3s of different clades and hosts. The black line represents the standard curve when the codon usage bias is determined by the GC3s composition only. The D/660, D/Japan and D/OK clades and swine and bovine hosts are represented in light blue, yellow, dark blue, grey and orange, respectively.
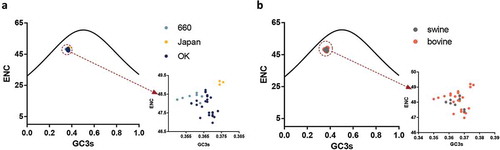
Figure 4. Neutrality plot analysis of GC3s against GC12s. a and b are diagrams of different clades and host, respectively. The IDV HEF strains cluster into three clades including: D/660, D/OK and D/Japan, represented in light blue, dark blue, and yellow, respectively. The line and dot of swine and bovine are represented in grey and orange, respectively.
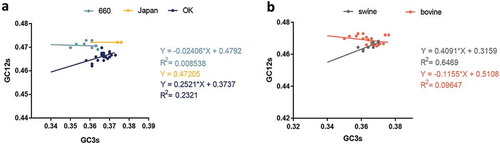
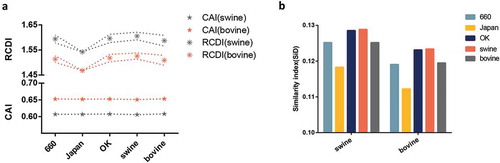
Figure 5. (a) CAI analysis (bottom panel represented by a symbol star) and RCDI analysis (upper panel represented by a symbol asterisk) of the HEF gene in relation to the natural hosts. The lines in the RCDI analysis in each host represent the upper and lower limit. (b) SiD analysis of the IDV HEF gene. The D/660, D/Japan, and D/OK clades are represented in light blue, yellow, dark blue. The swine, goat, and bovine hosts are represented in grey, dark green, and orange, respectively. The x axis represents the sequences belonging to different clades or identified in different hosts.