Figures & data
Table 1. Eicosanoids produced by human pathogenic yeasts and genes identified for PGE2 production in C. albicans, C. parapsilosis and C. neoformans.
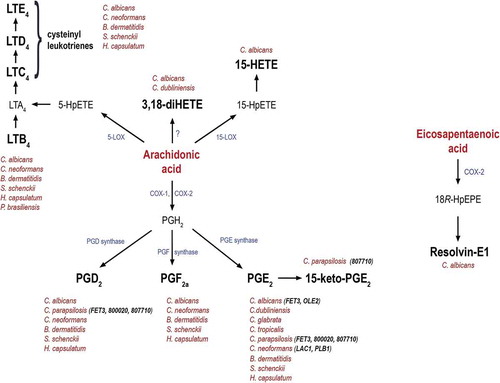
Figure 1. Schematic representation of eicosanoid biosynthesis, their production by fungi and the corresponding genes involved in their production.
Eicosanoid production from the precursor arachidonic acid or eicosapentaenoic acid. Besides mammals, cysteinyl leukotrienes, 3,18-diHETE, 15-HETE, PGD2, PGF2a, PGE2, 15-keto-PGE2 and resolvin-E1 are also produced by different pathogenic fungi. Although the exact biosynthetic pathways remain unknown, several fungal genes have been proposed to regulate their synthesis. These include FET3 and OLE2 in C. albicans, FET3 (CPAR2_603600), CPAR2_800020 and CPAR2_807710 in C. parapsilosis, and LAC1 and PLB1 in C. neoformans.