Figures & data
Figure 1. Sequences of Pex13 homologs and Pex14 homologs in fungal species. (a-b) The putative protein sequences of Pex13 and Pex14 homologs, Anpex13 (XP_659115) and Anpex14 (CBF76374.1) from Aspergillus nidulans, Copex13 (ENH77396.1) and Copex14 (ENH79683.1) from Colletotrichum orbiculare, Fgpex13 (XP_011316361) and Fgpex14 (XP_011328505.1) from Fusarium graminearum, Mopex13 (XP_003718970.1) and Mopex14 (XP_003717914.1) from Magnaporthe oryzae, Ncpex13 (XP_965021.3) and Ncpex14 (XP_957540.1) from Neurospora crassa, and Scpex13 (KZV09435.1) and Scpex14 (KZV11075.1) from Saccharomyces cerevisiae were retrieved and aligned using ClustalX. Identical amino acids are highlighted against a black background, conserved residues against a dark gray background, and similar residues against a light gray background. The putative conserved transmembrane domain 1 (TMD1) and 2 (TMD2) domain and SH3 domain in the Pex13 proteins are underlined in blue. The putative hydrophobic region and coiled-coil structure in the Pex14 proteins are underlined in red. The phylogenetic relationships of the Pex13 homologs (c) and the Pex14 homologs (d) respectively were calculated using the neighbor joining method in the MEGA 6.0 software.

Figure 2. Cellular localization of Mopex13 and Mopex14. (a) Mopex13 and Mopex14 are distributed on the peroxisomal membrane. Fluorescent microscopic analysis of the cotransformants GFP-Mopex13/mCherry-PTS1 and GFP-Mopex14/mCherry-PTS1 in Magnaporthe oryzae wild type Guy-11. GFP-Mopex13 and GFP-Mopex14 were both distributed in a punctate pattern, overlaying well with the peroxisome marker mCherry-PTS1. (b) The deletion of Mopex13 and Mopex14 did not alter the peroxisomal distribution of each other but led to a cytoplasmic distribution of Mopex14/17. The lack of Mopex14/17 did not change the distribution of Mopex13 or Mopex14. Bars = 5 µm.
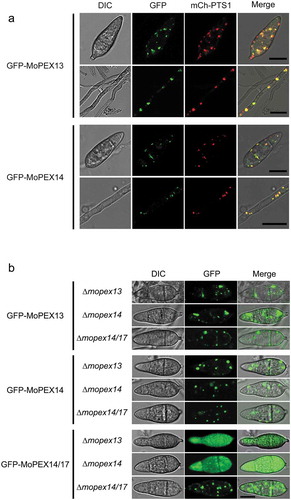
Figure 3. Interactions of Mopex13, Mopex14, Mopex14/17 and PTS receptors. The interactions between Mopex13, Mopex14 and Mopex14/17 (a), and the interactions of Mopex13, Mopex14 and Mopex14/17 with Mopex5 and Mopex7 (b) were analyzed by the yeast two-hybrid assay. The proteins were fused to the GAL4 DNA-binding domain (GAL4-BD) and the GAL4-activation domain (GAL4-AD), respectively. The indicated plasmid combinations were cotransformed into the Y2HGold yeast strain (Clontech). The transformants were tested for blue coloration on leucine/tryptophan double dropout plates presenting X-a-Gal and for viability on leucine/tryptophan/adenine/histidine quadruple dropout plates.
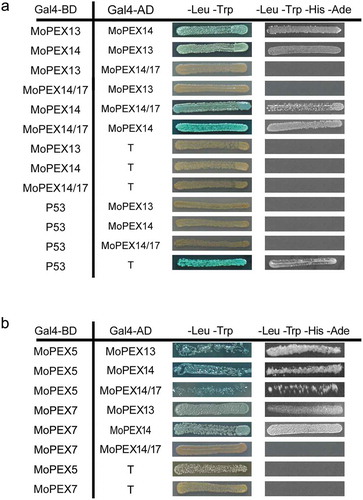
Figure 4. Mopex13 and Mopex14 are required for the distribution of peroxisomal matrix proteins and the Woronin body protein HEX1. The mCherry fused to PTS1 (mCh-PTS1), GFP fused to PTS2 (PTS2-GFP), Pmp47 labeled with GFP (GFP-Pmp47), HEX1 labeled with mCherry (mCh-HEX1), and Mopex5 labeled with GFP (GFP-MoPEX5) were respectively introduced into wild type Guy-11, the ∆mopex13 and ∆mopex14 mutants, as well as the ∆mopex14/17 mutants for GFP-MoPEX5. The peroxisomal distribution patterns of mCh-PTS1, PTS2-GFP, mCh-HEX1 were dramatically changed in the ∆mopex13 and ∆mopex14 mutants, while those of GFP-Pmp47 and GFP-MoPEX5 were unaltered compared to those in the wild type. The distribution of GFP-MoPEX5 was also unchanged in the ∆mopex14/17 mutant. Bars = 5 µm.
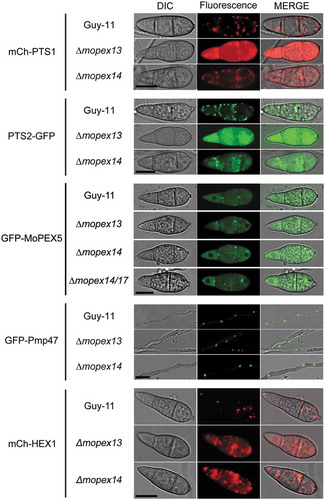
Figure 5. Ultrastructure of ∆mopex13 and ∆mopex14 mutants. Hyphae and conidia harvested from 5-day-old CM plates were analyzed by TEM. P, peroxisome; S, septum; Wbs, Woronin bodies; M, mitochondrion; N, nucleus; Sp, septal pore; WLS, Woronin body-like structure. Bars = 0.5 μm.
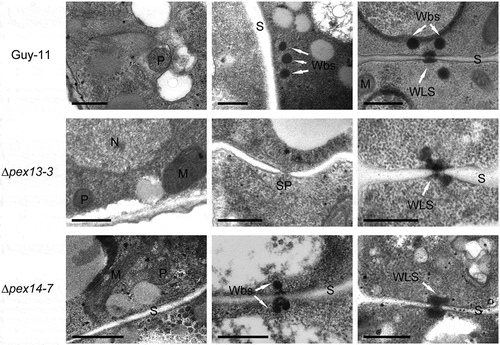
Figure 6. Vegetative growth, conidiation and melanization of the ∆mopex13 and ∆mopex14 mutants. The ∆mopex13 and ∆mopex14 mutants, wild type and complemented strains were cultured on 9-cm CM plates at 28°C for 10 days (a&b). The radical and aerial hyphal growth of the strains was compared (c). Conidiation of the strains was recoded and compared (d&e). Bar = 50 µm. The strains were cultured in CM liquids with 12-h light/12-h dark cycles at 28°C for 4 days, and then the mycelia were harvest and weighed (f). Melanization of the cultures was recorded and compared by measuring the absorbance value of the culture solutions at 415 nm (OD415) (g&h). Means and standard errors were calculated from three independent repeats. Single asterisks indicate significant differences at P < 0.05 compared with the wild-type, and double asterisks indicate significant differences at P < 0.01.
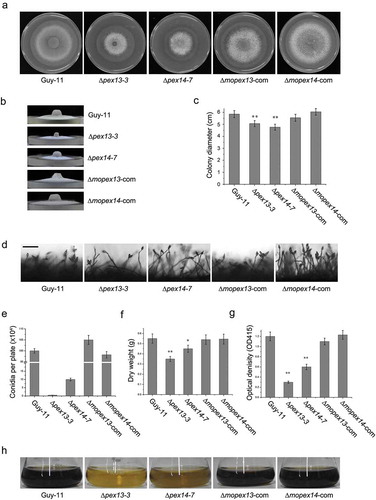
Figure 7. Mopex13 and Mopex14 are indispensable for pathogenicity on rice and barley. (a) Two-week-old rice cultivar CO39 seedlings were spray-inoculated with the conidial suspension (1 × 105 conidia/ml). The symptoms were recorded at 7 days postinoculation (dpi). (b) Seven-day-old barley cultivar ZJ-8 seedlings were spray-inoculated with the conidial suspension (1 × 105 conidia/ml) and recorded at 3 dpi. (c) Detached barley leaves were inoculated with 20 μl of the conidial suspensions (upper, 1 × 105 conidia/ml; lower, 1 × 104 conidia/ml) and incubated for 4 days. (d) Droplet-inoculated barley leaves were sampled at 24, 36 and 48 hpi, discolored, and examined under a light microscope to detect the infectious structures. Bar = 20 µm.

Figure 8. Mopex13 and Mopex14 are required in appressorial development and host penetration. The conidia of ∆mopex13, ∆mopex14 and wild type harvested from 9-day-cultured CM plates and suspended at 1 × 105 conidia/ml were incubated to allow germination and appressorial formation. (a&b) The conidial germination rates and appressorial formation rates at sequential time points were calculated and statistically compared. (c) Microscopy of the appressorial development of the strains. (d&e) Incipient-cytorrhysis was performed to compare the appressorial turgor genesis in the 24-h appressoria of strains using glycerol or PEG8000 as the external solute. Means and standard errors were calculated from three independent replicates (at least 100 conidia each). (f) The relative transcription of MoPEX13 and MoPEX14 during appressorial development was examined by qRT-PCR using the tubulin gene as a reference. (g) Inoculation of cuticle-removed (wound) barley leaves with 1 × 105 conidia/ml conidial suspensions. The mutants partially regained the ability to cause lesions on wounded leaves. (h) Infectious structure on inoculated intact and wounded barley leaves.
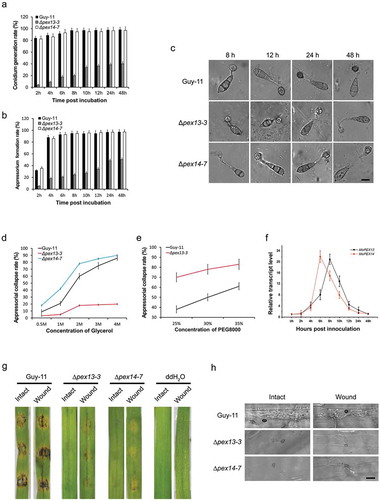
Figure 9. Mopex13 and Mopex14 are involved in lipid metabolism. (a) Lipid utilization by ∆mopex13 and ∆mopex14 and the wild-type strain Guy-11. The strains were cultured on minimal medium with glucose (1%), Tween 80 (0.5%), olive oil (0.5%) or sodium acetate (50 mM) as the sole carbon source for 5 days. (b) Relative transcription of MoPEX13 and MoPEX14 in the wild type cultured on minimal medium with glucose (1%) or with glucose (0.5%) together with Triolein (0.5%), Tween 80 (0.5%), olive oil (0.5%) or sodium acetate (50 mM). (c) Deletion of MoPEX13 or MoPEX14 delayed lipid mobilization and degradation during appressorium development. Conidia of the wild-type strain Guy-11 and mutants were incubated on a hydrophobic membrane and allowed to form appressoria. Samples at different time points were stained with Nile Red and examined microscopically. Bar = 5 μm. (d) Inoculation of barley leaves with conidial suspensions (1 × 105 conidia/ml) supplemented with glucose (2.5%). The presence of glucose partially improved the ability of the mutants to cause disease.
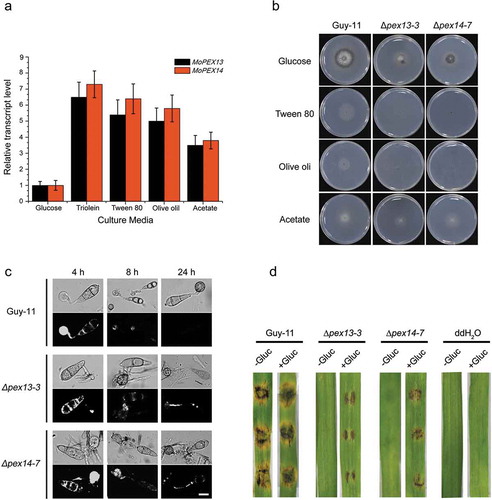
Figure 10. Deletion of MoPEX13 or MoPEX14 reduced the tolerance of the mutants to ROS. (a) The ∆mopex13, ∆mopex14 and wild type strains were cultured on complete medium in the presence of hydrogen peroxide (0.04%, v/v), methyl viologen (MV, 0.25 mg/ml) or Rose Bengal (RB, 100 μM). Olive oil (0.5%) or sodium acetate (50 mM) was used as the sole carbon source for 7 days. (b-d) The colony diameters of the tested strains were measured to calculate and compare significant differences in habitation rates. Means and standard errors were calculated from three independent replicates. Single asterisks indicate significant differences at P < 0.05 compared with the wild-type, and double asterisks indicate significant differences at P < 0.01. (e) Inoculation on barley leaves that were heat-treated to suppress ROS generation. The heat treatment improved the pathogenicity of the mutants on the barley leaves.
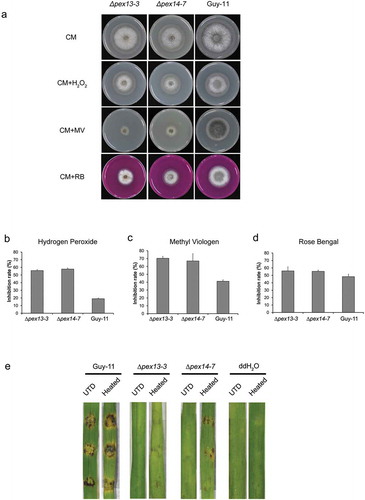
Figure 11. Deletion of MoPEX13 or MoPEX14 reduced the tolerance of the mutants to cell wall interference agents. (a) The ∆mopex13, ∆mopex14 and wild type strain were cultured on CM supplemented with Congo red (100 µg/ml) or Calcofluor white (50 µM) for 7 days. (b&c) Statistical comparison of the radial growth of the strains. Error bars represent the deviation from three replicates, and double asterisks indicate significant differences in comparison to the wild type at P < 0.01.
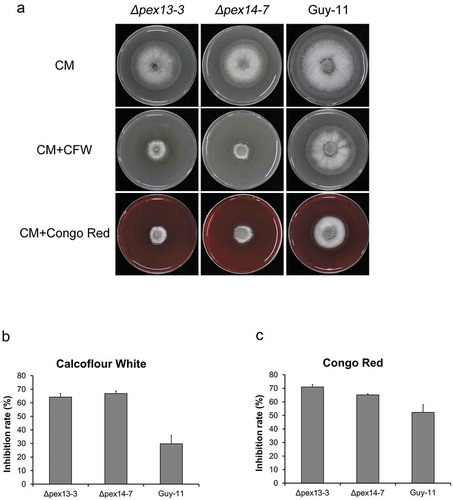
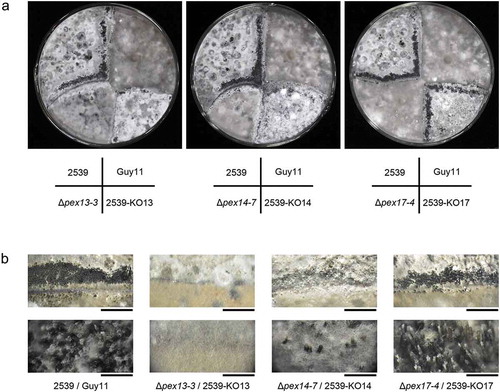
Figure 12. Mopex13 and Mopex14 are required in sexual reproduction of M. oryzae. The ∆mopex13, ∆mopex14 and ∆mopex14/17 mutants derived from Guy-11 strain and those from 2539 strain were cross-cultured on oatmeal agar plates at 22°C for 30 days. (a) The asci formed at the junction of the strains were recorded. (b) Magnified images of the junction areas of the strains. Bars in the upper panel = 5 mm; bars in the lower panel = 1000 μm.