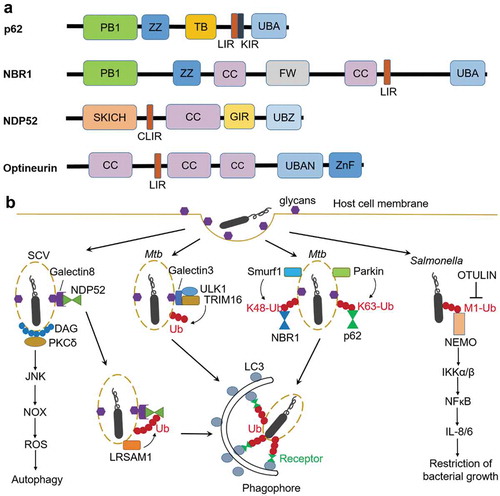
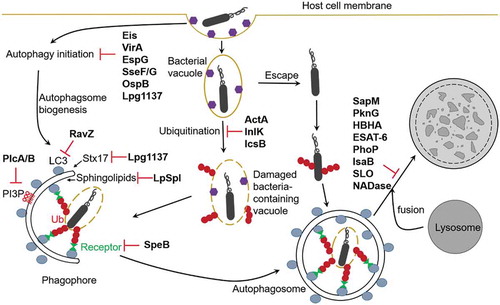
Figures & data
Table 1. Mechanisms involved in the interaction of intracellular bacteria with host autophagy.