Figures & data
Figure 1. Phylogenetic relationships for Clostridiales species based of the concatenated sequence of 40 high-resolution molecular markers. a) Inter-species phylogenetic relationships for the selected genome dataset. A set of 65 Clostridiales genomes, belonging to eight families, was analyzed to propose the taxonomic designation of the target genomes (C. tertium and C. paraputrificum). Three representative genomes per family were included except for the Heliobacteriaceae family, where a single genome passed the preliminary quality tests, and for the Clostridiaceae family, for which a per-species genome was sought, because it was the family of interest. In the external ring, color was assigned to each Clostridiales family that was used in each analysis. The target genomes are marked with gray boxes, while boxes with dotted lines mark the grouping node for each target species. The Bacillus coagulans strain HM-08 (GCF_000876545.1) genome was included as outgroup. Red dots represent bootstrap values of ≥ 90.0.
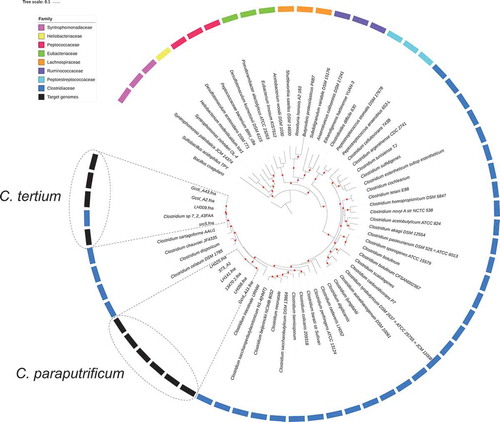
Table 1. Genome features of the C. tertium and C. paraputrificum clinical isolates.
Table 2. Groups used for the intra-taxa comparisons.
Figure 2. Intra-taxa comparisons of the complete genome sequences for (a) C. tertium and (b) C. paraputrificum. Analyses were developed in the CGview server [Citation57]. The genome with the biggest size was selected as the reference for each species. Regions in white correspond to variable areas between genomes. Colors were assigned to the outer rings when sequences were found that each had a BLAST identity cutoff of 0.6, for the pairwise comparisons against the “reference genome”.
![Figure 2. Intra-taxa comparisons of the complete genome sequences for (a) C. tertium and (b) C. paraputrificum. Analyses were developed in the CGview server [Citation57]. The genome with the biggest size was selected as the reference for each species. Regions in white correspond to variable areas between genomes. Colors were assigned to the outer rings when sequences were found that each had a BLAST identity cutoff of 0.6, for the pairwise comparisons against the “reference genome”.](/cms/asset/cb0bb6f2-a965-427a-b50e-71574d86d904/kvir_a_1637699_f0002_oc.jpg)
Figure 3. Pan-genome analyses for C. tertium (a) and C. paraputrificum (b) species as determined by Roary [Citation61]. The parameters were defined as follows: Core genes: 99% ≤ strains ≤ 100% -accessory genes (15% ≤ strains < 95%). evolutionary insights between isolates based on the core genome (left panel). The core-genome tree generated was compared with a matrix where the core and accessory genes were either present or absent, which was graphically represented using the ipython roary_plots.py script. The top panel shows the color assignments for the different genes according to their lengths. NAG: number of accessory genes; NUG: number of unique genes. Red dots represent bootstrap values of ≥ 99.0.
![Figure 3. Pan-genome analyses for C. tertium (a) and C. paraputrificum (b) species as determined by Roary [Citation61]. The parameters were defined as follows: Core genes: 99% ≤ strains ≤ 100% -accessory genes (15% ≤ strains < 95%). evolutionary insights between isolates based on the core genome (left panel). The core-genome tree generated was compared with a matrix where the core and accessory genes were either present or absent, which was graphically represented using the ipython roary_plots.py script. The top panel shows the color assignments for the different genes according to their lengths. NAG: number of accessory genes; NUG: number of unique genes. Red dots represent bootstrap values of ≥ 99.0.](/cms/asset/48318ea0-e68b-4ac9-8c9e-e5f0d931e42a/kvir_a_1637699_f0003_oc.jpg)
Table 3. Virulence factors and antimicrobial resistance-related molecular markers identified across C. tertium and C. paraputrificum genomes.
Figure 4. Identification of the proteins involved in sporulation processes for each analyzed species. As reference the reported proteins in C. botulinum Ba4 657 uid59173 were used as comparisons in the CD-HIT suite, taking the following parameters into account: % identity > 40 and Kmer = 2. * Stage: sporulation pathway step based on the reported information. ** Cbot: C. botulinum Ba4 657 uid59173; Numbers inside each well indicate the reference genome length and the numbers in parenthesis represent the number of copies found. Parenthetical numbers indicate the number of times proteins were identified more than once in the multifasta files that were used for each analysis. † Black boxes represent the proteins reported (in the reference genome), that are missing in all the evaluated isolates. Because no sporulation proteins were identified in 72_43FAA (C. tertium) and LH058 (C. paraputrificum) they were excluded from the analyses. The proteins are listed according to their timing of action in the sporulation pathways, in agreement with previous reports [Citation78,Citation98].
![Figure 4. Identification of the proteins involved in sporulation processes for each analyzed species. As reference the reported proteins in C. botulinum Ba4 657 uid59173 were used as comparisons in the CD-HIT suite, taking the following parameters into account: % identity > 40 and Kmer = 2. * Stage: sporulation pathway step based on the reported information. ** Cbot: C. botulinum Ba4 657 uid59173; Numbers inside each well indicate the reference genome length and the numbers in parenthesis represent the number of copies found. Parenthetical numbers indicate the number of times proteins were identified more than once in the multifasta files that were used for each analysis. † Black boxes represent the proteins reported (in the reference genome), that are missing in all the evaluated isolates. Because no sporulation proteins were identified in 72_43FAA (C. tertium) and LH058 (C. paraputrificum) they were excluded from the analyses. The proteins are listed according to their timing of action in the sporulation pathways, in agreement with previous reports [Citation78,Citation98].](/cms/asset/a74f41a6-63df-42c7-8f99-73a49f3d20d1/kvir_a_1637699_f0004_oc.jpg)
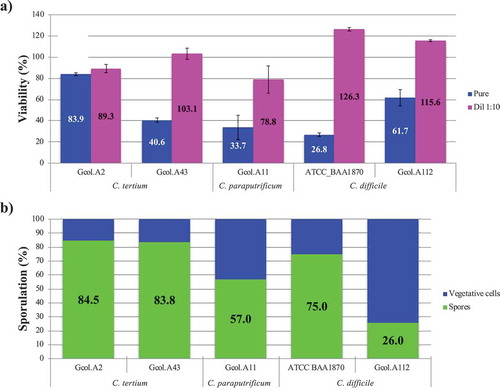
Figure 5. Experimental validation of the cytotoxic effect (a) and the sporulation capacity (b) of each Colombian isolate. C. difficile isolates considered to be toxigenic (ATCC_BAA1870) and non-toxigenic (Gcol.A112) were included as controls. The percentage viability (%), as measured by the MTT assay, is expressed in comparison with the negative control, where DMEM medium was used to replace the volume of the isolate with 50% (v/v) saline solution, after which all the samples received the same experimental treatment. Each isolate supernatant was evaluated directly and as a 1:10 solution. The sporulation percentage, as measured by flow cytometry, was based on the size (≤0.3 μM) and granularity (≤0.9 μM). The percentage sporulation was calculated by considering the number of spores with respect to the total number of particles analysed.