Figures & data
Figure 1. Phylogenetic tree analysis and domain prediction among casein kinase homologs. (a). The phylogenetic tree was constructed by MEGA 5.0 using the neighbor-joining tree construction method with 1000 bootstrap replicates. Sequences of casein kinase homologs were obtained from NCBI databases and aligned by Clustal Omega. Accession numbers of homologs are indicated in the figure. (b). Domains contained in casein kinases were predicted using the SMART webserver. The picture was drawn using DOG 2.0 software.
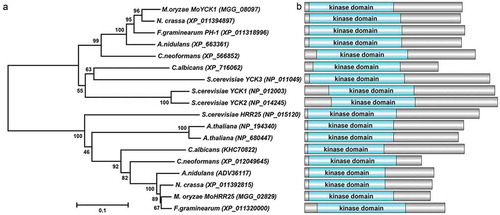
Figure 2. MoYck1 is involved in growth and sexual reproduction in M. oryzae. (a). Growth of Guy11, ΔMoyck1 and Moyck1c on CM, V8, and OMA plates. Mycelial plugs were inoculated on the above plates for 8 days before photography. (b). Diameters of colonies were measured and mean and standard deviation were presented calculated from data of three replicates. The same characteristic showed no significant differences (Duncan’s test, P < 0.01), and the error bars represent the standard deviation. (c). Utilization of different carbon sources. Mycelial plugs of Guy11, ΔMoyck1 and Moyck1c were inoculated on MM plates with glucose, sucrose, and mannose for 7 days before photography. (d). Relative growth ratios of Guy11, ΔMoyck1 and Moyck1c. The averages are calculated from measuring three replicates, representing relative ratios of growth on sucrose and mannose relative to that on glucose. The data is subject to Duncan’s test and a significant difference is shown in the figure (P < 0.05). (e). Sexual development. Crossing of Guy11 or ΔMoyck1 backcrossed with the opposite mating strain 2539 under constant fluorescent light at 20°C to induce sexual development. Arrows indicate the formation of perithecia at the junction of Guy11 and 2539.
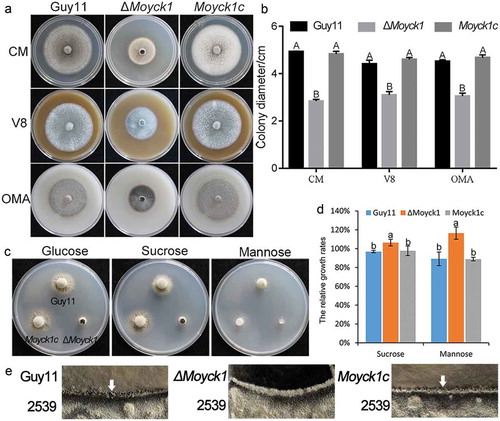
Figure 3. MoYck1 is required for conidiogenesis and conidial morphology. (a). Observation of conidiophore differentiation. Bar = 100 µm. (b). Conidiation was measured at 7 dpi on CM plates. C. CFW staining. Conidia from Guy11, ΔMoyck1 and Moyck1c were stained by CFW. (d). Conidial septation. The proportions of conidia with various numbers of septa were counted. (e). Size of conidia. The length and width of more than two hundred conidia were measured. Different letters indicate a significant difference by Duncan’s test (P < 0.01), with error bars representing the standard deviation.
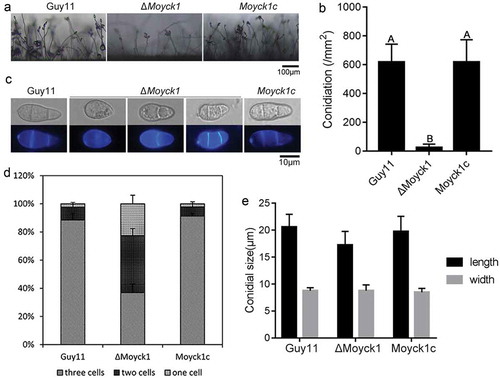
Figure 4. MoYck1 is responsible for conidial germination and appressorium formation. (a). Conidial germination was observed at 4 and 24 hours post incubation on the hydrophobic surface. (b). Ratios of germination and appressorium formation were counted. The data with the same characters indicate no significant differences (Duncan’s test, P < 0.01).
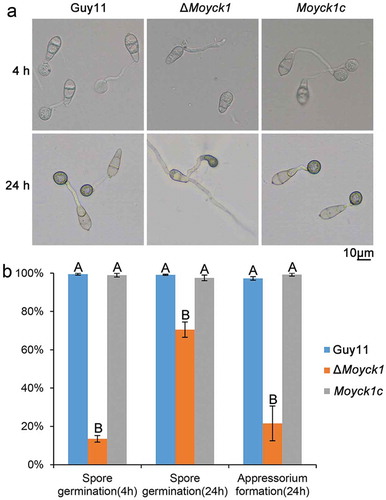
Figure 5. MoYck1 contributes to full virulence. (a). Virulence on rice seedlings. Conidia (1 × 104/mL) from Guy11, ΔMoyck1 and Moyck1c were sprayed on 2-week-old rice seedlings, and pictures were taken at 5 dpi. (b). Virulence on barley leaves. Conidial drops were inoculated on barley leaves in vitro. Photos were taken at 5 dpi. (c). Appressorium-mediated penetration on barley leaves. The barley leaves were inoculated with conidia for 48 hours and then decolored before observation. Bar = 20 µm. (d). Ratios of appressorium-mediated penetration. The values mean average and bars mean standard errors. The column labeled with different characters means an apparent difference (Duncan’s test, P < 0.01).
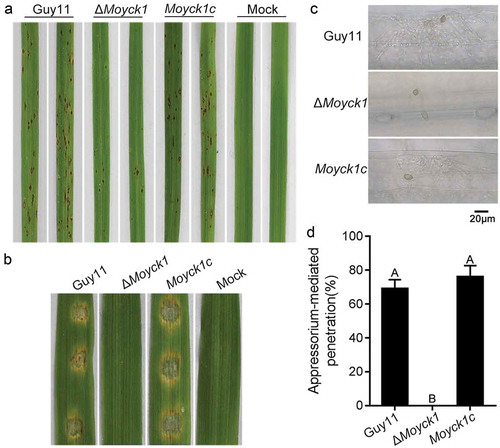
Figure 6. Autophagy is negatively regulated by MoYck1 in M. oryzae. (a). Observation of autophagic affluxes in Guy11 and ΔMoyck1. The degradation of GFP-MoAtg8 was observed via western blotting with an anti-GFP antibody. GAPDH was used to indicate the loading amount of total protein. (b). Lipidation of MoAtg8 observed in M. oryzae. Lipidation of MoAtg8 was observed in ΔMoatg4, ΔMoatg3 and Guy11 under nitrogen starvation conditions for 3 and 6 hours via western blotting with anti-MoAtg8. (c). Lipidation of MoAtg8 compared with that in Guy11 and ΔMoyck1. Lipidation of MoAtg8 and the amount of MoAtg8 were observed in Guy11 and ΔMoyck1. The protein GAPDH was used as a loading control.
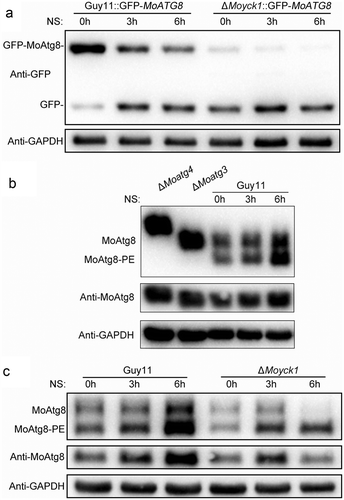
Figure 7. MoVps41 positively regulates autophagic processes. (a). Colocalization of MoVps41 and MoAtg8. GFP-labeled MoVps41 and DsRed-labeled MoAtg8 were observed at the hyphal and conidial stages. Bar = 5 µm. (b). Protein degradation assay of GFP-MoAtg8 by western blotting in Guy11 and ΔMovps41. Total proteins of Guy11 and ΔMovps41 expressing the GFP-MoAtg8 fusion protein were extracted from mycelia at the indicated time after nitrogen starvation and were detected with anti-GFP antibody. (c). Microscopic observation of GFP-MoAtg8 localization. After nitrogen starvation treatment, GFP-MoAtg8 entered the vacuoles and colocalized with the CMAC-labeled vacuole in Guy11. However, GFP-MoAtg8 in ΔMovps41 still displayed dotted fluorescence outside the vacuoles. Bar = 2 µm. (d). Lipidation of MoAtg8 in Guy11 and ΔMovps41. Lipidation of MoAtg8 was induced with nitrogen starvation for 4 hours and detected via western blotting.
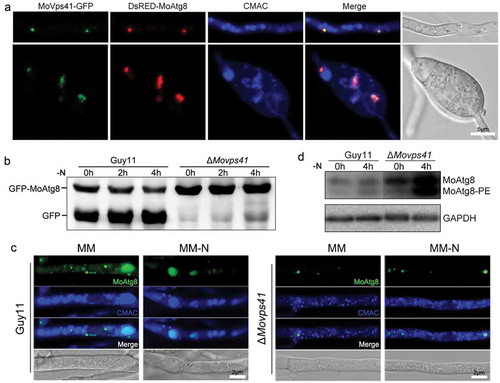
Figure 8. MoYck1 is involved in the osmotic stresses response via regulating the phosphorylation of Osm1. (a). Cultures of Guy11, ΔMoyck1 and the complemented strain Moyck1c on media with hyperosmotic reagents including 0.5 M NaCl, 0.7 M KCl, 1 M sorbitol and 1 M sucrose. Pictures were taken at 7 dpi. (b). The relative inhibition rates of Guy11, ΔMoyck1 and the complemented strain Moyck1c on media with hyperosmotic reagents. Each strain was cultured for three replicates. The columns indicate average values, and the error bars indicate standard deviation. The same letters indicate no apparent differences (Duncan’s test, P < 0.01). (c). Phosphorylation of Osm1. The phosphorylation of Osm1 in Guy11 and ΔMoyck1 was detected following induction with 0.5 M NaCl at 0, 10, 30, 60, and 120 min. The protein GAPDH was used as a loading control. (d). The relative content of phosphorylated Osm1. The amounts of phosphorylated Osm1 in Guy11 and ΔMoyck1 were compared with those of GAPDH at the indicated time points.
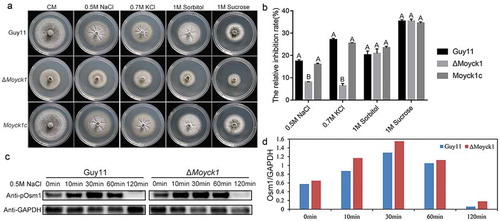
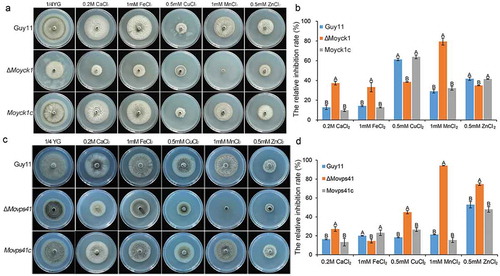
Figure 9. MoYck1 and MoVps41 play crucial roles in the detoxification of heavy metal ions. (a). Mycelial plugs of Guy11, ΔMoyck1 and the complemented strain Moyck1c were inoculated onto 1/4 YG plates with different metal ions. Images were obtained at 6 dpi. (b). The growth inhibition rates were calculated in three independent assays with three replicates each time. Columns labeled with different letters indicate a significant difference determined by Duncan’s test (P < 0.01). (c). Mycelial plugs of Guy11, ΔMovps41 and the complemented strain Movps41c were inoculated onto 1/4 YG plates with different metal ions. Images were obtained at 6 dpi. (d). The growth inhibition rates of Guy11, ΔMovps41 and the complemented strain Movps41c were assessed in three independent assays with three replicates. Columns labeled with different letters indicate a significant difference determined by Duncan’s test (P < 0.01).