Figures & data
Figure 1. Genetic organization and domain structure of OI-9 in O157. (a) Graphic representation of the region surrounding OI-9 in the genome of O157 EDL933 segment 323657–330128. Arrows represent open reading frames. (b) Domain structure of z0342, z0343, z0346, z0347, and z0348 in OI-9.
Figure 2. Adherence of ΔovrB mutants in vitro. (a,b) Adherence of O157 WT, ΔovrB mutant, and ovrB complementary strain to HeLa (a) and Caco-2 (b) cells. (c) Growth of O157 WT, ΔovrB mutant, and ovrB complementary strain in LB medium. (d) Detection of AE lesion formation by O157 WT, ΔovrB mutant, and ovrB complementary strain by FAS in HeLa cells at 3 h. The HeLa cell actin cytoskeleton (green) and nuclei of bacterial and HeLa cells (red) are shown. AE lesions are indicated by arrowheads. (e) The number of pedestals/infected HeLa cells by O157 WT, ΔovrB mutant, and ovrB complementary strain (n = 150 cells). (f) qRT-PCR analysis of changes in LEE gene expression in O157 WT, ΔovrB mutant, and the ovrB complementary strain. Data represent mean ± SD (n = 3). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (Student’s t-test).
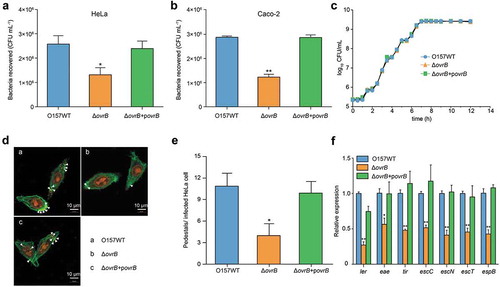
Figure 3. Ler mediates activation of LEE genes by OvrB in O157. (a-e) EMSA of the specific binding of OvrB to PLEE1 (a) rpoS (negative control) (b), PLEE2/3 (c), PLEE4 (d), and PLEE5 (e). PCR products were added to the reaction mixtures at 40 ng each. OvrB protein was added to the reaction buffer in lanes 2–7 at 0.1, 0.2, 0.4, 0.8, 1, and 2 μM, respectively. No protein was added in lane 1. (f) Fold enrichment of the LEE1, LEE2/3, LEE4, and LEE5 promoters in OvrB ChIP samples, as measured by qPCR. rpoS served as negative control. (g) Adherence of O157 WT, ΔLer mutant, and ΔLerΔovrB double mutant to HeLa cells. (h) qRT-PCR analysis of changes in LEE gene expression in O157, ΔLer mutant, and ΔLerΔovrB double mutant. Data represent mean ± SD (n = 3). *P ≤ 0.05; **P ≤ 0.01, ***P ≤ 0.001 (Student’s t-test).
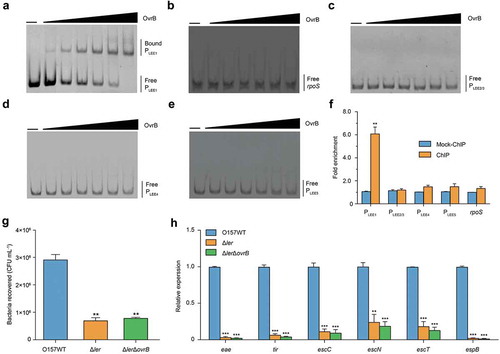
Figure 4. Adherence capacity of O157 in the mouse intestinal tract. (a-c) Evaluation of the adherence capacity of O157 WT, ΔOI-9 mutant, ΔovrB mutant, and ΔovrB complementary strain in distal colon of mice at 6 h (a), 2 d (b), and 4 d (c) post-infection. Horizontal lines represent the geometric means. Statistical significance was assessed with the Mann-Whitney rank-sum test.
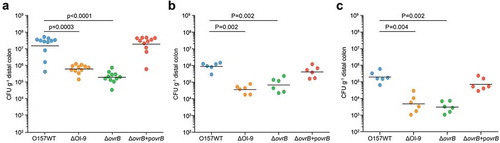
Figure 5. OvrB is a ubiquitous regulator of virulence in pathogenic bacteria. (a) Adherence of G2583 (E. coli O55:H7), G1345 (E. coli O145:H28), and orthologous ΔovrB mutant to HeLa cells. (b,c) qRT-PCR analysis of changes in LEE gene expression in G2583 (E. coli O55:H7) and the orthologous ΔovrB mutant (b) and in G1345 (E. coli O145:H28) and the orthologous ΔovrB mutant (c). Data represent mean ± SD (n = 3). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (Student’s t-test).
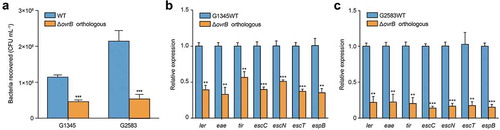
Figure 6. Model of the OvrB regulatory network in O157. Scheme of OvrB regulation of virulence-associated LEE genes. OvrB directly activates LEE1 by binding to the promoter region, which in turn increases the transcript level of ler, allowing the Ler protein to function as a downstream activator. Broken black arrows correspond to translated products from these genes. Blue arrows represent input points of positive transcriptional regulation.