Figures & data
Table 1. Primers used in this study
Figure 1. Design and validation of primers specific for five different variants of tet(X) [X1 to X5] (a). Cartoon scheme for physical locations of five pairs of tet(X) primers. The rectangle represents a given tet(X) gene, and an arrow denotes the primer (b). Use of routine PCR to verify the feasibility of tet(X) primers. The PCR products were separated by an electrophoresis with 2.0% agarose gel. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol of “–”, negative control
![Figure 1. Design and validation of primers specific for five different variants of tet(X) [X1 to X5] (a). Cartoon scheme for physical locations of five pairs of tet(X) primers. The rectangle represents a given tet(X) gene, and an arrow denotes the primer (b). Use of routine PCR to verify the feasibility of tet(X) primers. The PCR products were separated by an electrophoresis with 2.0% agarose gel. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol of “–”, negative control](/cms/asset/9eb7442c-14af-4e84-b091-23557262ab24/kvir_a_1706913_f0001_oc.jpg)
Table 2. Field and clinical isolates examined in this study
Figure 2. Evaluation of specificity of the five sets of tet(X) [X1 to X5] primers (a). PCR assay suggests that the pair of primer [tet(X1)-F plus tet(X1)-R] is exclusively specific for tet(X1)-containing sample (b). The specificity of the primer [tet(X2)-F plus tet(X2)-R] is validated with PCR assays supplemented with the template of a single colony carrying different tet(X) gene (c). Among five kinds of different samples, only tet(X3)-bearing one is positive in PCR detection with the specific primers of tet(X3)-F plus tet(X3)-R (d). The tet(X4)-harboring sample is exclusively recognized by a single PCR with a pair of primers [tet(X4)-F plus tet(X4)-R] (e). The specificity of the primers [tet(X5)-F plus tet(X5)-R] is unique to the tet(X5)-positive sample PCR products were separated with electrophoresis of 2% agarose gel. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol of “–”, negative control
![Figure 2. Evaluation of specificity of the five sets of tet(X) [X1 to X5] primers (a). PCR assay suggests that the pair of primer [tet(X1)-F plus tet(X1)-R] is exclusively specific for tet(X1)-containing sample (b). The specificity of the primer [tet(X2)-F plus tet(X2)-R] is validated with PCR assays supplemented with the template of a single colony carrying different tet(X) gene (c). Among five kinds of different samples, only tet(X3)-bearing one is positive in PCR detection with the specific primers of tet(X3)-F plus tet(X3)-R (d). The tet(X4)-harboring sample is exclusively recognized by a single PCR with a pair of primers [tet(X4)-F plus tet(X4)-R] (e). The specificity of the primers [tet(X5)-F plus tet(X5)-R] is unique to the tet(X5)-positive sample PCR products were separated with electrophoresis of 2% agarose gel. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol of “–”, negative control](/cms/asset/49ada51e-6d4e-43d8-a387-1dbf130835a6/kvir_a_1706913_f0002_oc.jpg)
Figure 3. Establishment of different multiplex PCR assays for the tet(X) family genes (a). Development of multiplex PCR approaches to detect different combinations of double tet(X) genes in a single colony (b). Harnessing multiplex PCR approaches for detection of three tet(X) genes in a single colony (c). Developing efficient multiplex PCR assays to detect quadruple and even quintuple variants of tet(X) from a single colony. The specific PCR products with known sizes in a single PCR act as references. Namely, it denotes 486 bp for tet(X1), 343 bp for tet(X2), 685 bp for tet(X3), 204 bp for tet(X4), and 265 bp for tet(X5) (), respectively. Two percent of agarose gel was applied to separate the mixture of PCR products. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol “–”, negative control
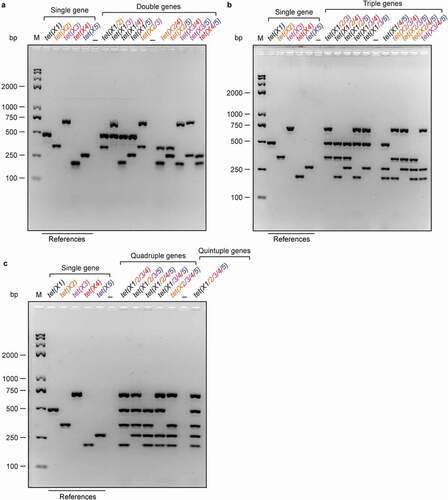
Figure 4. The use of multiplex PCR to screen tet(X) variants in field/clinical isolates. The stains examined here include field isolates and clinical isolates. All the field isolates were provided by the collaborator Dr. Jian Sun from South China Agriculture University, Guangzhou, China (). Among them, 17 are detected to be positive for tet(X) variants, including 8 tet(X5)-positive Acinetobacter isolates (Q477-2, Q478-2, Q479-2, Q480-2, CQW11-1, CQW13-2, CQS29-1 & CQS35-1), and 7 tetX4-bearing E. coli isolates (LHM10-1, G3X16-2, YSP8-1, SS13-1-1, SSS2-2, SSS3-1 & WF90-1), and 2 Acinetobacter isolates coharboring tet(X3) and tet(X5) (). The remaining eight strains (KP-1 to KP-8) refer to clinical isolates of Klebsiella pneumoniae with tigecycline resistance, collected from The Second Affiliated Hospital, Zhejiang University School of Medicine. Electrophoresis of 2% agarose gel was applied to separate the mixture of PCR products. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol “–”, negative control; the mixture of tet(X) [X1 to X5] PCR products, positive control
![Figure 4. The use of multiplex PCR to screen tet(X) variants in field/clinical isolates. The stains examined here include field isolates and clinical isolates. All the field isolates were provided by the collaborator Dr. Jian Sun from South China Agriculture University, Guangzhou, China (Table 2). Among them, 17 are detected to be positive for tet(X) variants, including 8 tet(X5)-positive Acinetobacter isolates (Q477-2, Q478-2, Q479-2, Q480-2, CQW11-1, CQW13-2, CQS29-1 & CQS35-1), and 7 tetX4-bearing E. coli isolates (LHM10-1, G3X16-2, YSP8-1, SS13-1-1, SSS2-2, SSS3-1 & WF90-1), and 2 Acinetobacter isolates coharboring tet(X3) and tet(X5) (Table 2). The remaining eight strains (KP-1 to KP-8) refer to clinical isolates of Klebsiella pneumoniae with tigecycline resistance, collected from The Second Affiliated Hospital, Zhejiang University School of Medicine. Electrophoresis of 2% agarose gel was applied to separate the mixture of PCR products. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol “–”, negative control; the mixture of tet(X) [X1 to X5] PCR products, positive control](/cms/asset/98506d5b-b194-41c1-b662-71c29a4b9112/kvir_a_1706913_f0004_oc.jpg)
Figure 5. Use of single PCR to re-verify the co-carriage of tet(X3) and tet(X5) in two isolates of CMG11-2 and FS35-1 PCR results of single gene verified that the two strains (CMG11-2 and FS35-1) are consistently positive in tet(X3) [and tet(X5)]-specific molecular detection, whereas negative in tet(X1) [or tet(X2)/tet(X4)]-specific PCR assays. The expected size of PCR products is 685 bp for tet(X3), and 265 bp for tet(X5) (). PCR products were separated with electrophoresis of 2% agarose gel. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol “–”, negative control; the mixture of tet(X) [X1 to X5] PCR products, positive control
![Figure 5. Use of single PCR to re-verify the co-carriage of tet(X3) and tet(X5) in two isolates of CMG11-2 and FS35-1 PCR results of single gene verified that the two strains (CMG11-2 and FS35-1) are consistently positive in tet(X3) [and tet(X5)]-specific molecular detection, whereas negative in tet(X1) [or tet(X2)/tet(X4)]-specific PCR assays. The expected size of PCR products is 685 bp for tet(X3), and 265 bp for tet(X5) (Table 1). PCR products were separated with electrophoresis of 2% agarose gel. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol “–”, negative control; the mixture of tet(X) [X1 to X5] PCR products, positive control](/cms/asset/40e85da6-17f3-4418-89bd-0efc994fe0ed/kvir_a_1706913_f0005_oc.jpg)
