Figures & data
Table 1. S. maltophiia strains used in this study and their average zone of twitching under normal and iron-depleted conditions
Table 2. Functional roles of type IV pilus associated with twitching motility and their abbreviations obtained from RAST server
Figure 1. (a) Typical subsurface twitching motility agar interpretation. A colony on the surface of agar around the inoculation point (top colony) and a visible halo or hazy zone of bacteria that have twitched across the plate between the bottom of the agar and the Petri plate (interstitial colony). (b) A representative of the zone of twitching motility on twitching agar plates on normal and iron-depleted conditions. P. aeruginosa ATCC27853 (positive control), SM45 (clinical isolate) and LMG10879 (environmental isolate). The plates were flooded with TM developer solution. The zone of twitching motility was marked, and 1 cm line was drawn onto the plates. (c) Box-and-whisker plot for clinical isolates under normal and iron-depleted conditions. The twitching zone under normal condition (min: 13 mm, median: 22 mm, max: 45 mm), while iron-depleted condition (min:14 mm, median: 25 mm, max: 50 mm)
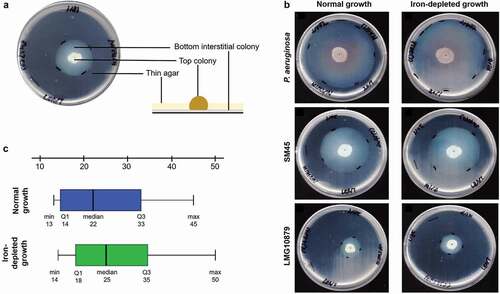
Table 3. Differentially expressed motility-associated proteins identified via ITRAQ assay for CS17 and LMG959 isolates under iron-depleted condition
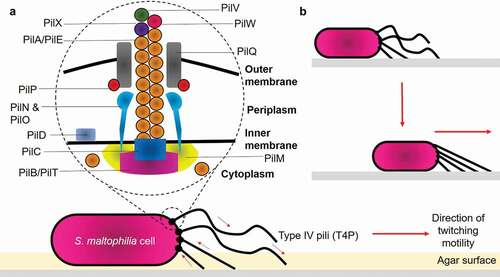
Figure 2. Structure of type IV pili and mechanisms of bacterial twitching motility. (a) Structurally, T4P is composed of: (1) Major pilin PilA/PilE; (2) Secretin PilQ; (3) Alignment proteins PilM, PilN, PilO, PilP; (4) Retraction ATPase PilT; (5) Assembly ATPase PilB; and (6) Minor pilins PilX, PilW, PilV. (b) Pilus retraction results in the forward movement of the cell across the surface. Arrows indicate the direction of pilus retraction/extension
Data availability statement
The datasets generated and/or analysed during the current study are available in the ProteomeXchange Consortium database (http://proteomecentral.proteomexchange.org) via PRoteomics IDEntifications (PRIDE) with the dataset identifier PXD004370.
