Figures & data
Table 1. Strains and plasmids used in the study
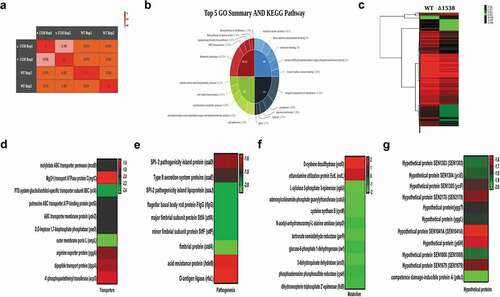
Figure 1. SEN1538 belongs to KGG superfamily of proteins. (a) Pairwise sequence alignment of STM1513 protein (GenBank Accession ID: AAL20432.1) from Salmonella enterica subspecies I serovar Typhimurium str. LT2 and SEN1538 protein (GenBank Accession ID: CAR33117.1) from Salmonella enterica subspecies I serovar Enteritidis str. P125109. (b) Superfamily and conserved domain of SEN1538 protein was screened by using NCBI Conserved Domain Tool and InterProScan5 Sequence Search Tool. (c) Multiple sequence alignment of KGG superfamily of proteins from different bacterial species against SEN1538 protein. Salmonella Enteritidis; Klebsiella pneumoniae, Salmonella Typhimurium; Salmonella Newport; Salmonella Gallinarum; Micromonospora sp. Rc5, Salmonella Typhi; Escherichia coli; Vibrio parahaemolyticus; Citrobacter amalonaticus.
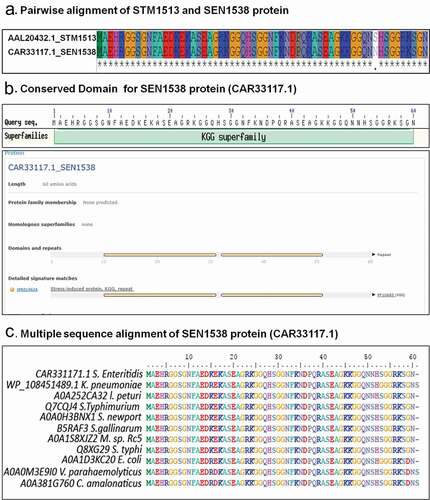
Figure 2. SEN1538 promotes resistance to stressors. The Log-phase cultures of Δ1538 and the complemented strain cΔ1538 were assessed under different stress conditions in comparison to WT (a) Antimicrobial peptides (AMPs) stress survival assay (b) Expression of SEN1538 in WT after treatment with AMPs through qRT-PCR. (c) Expression of AMP resistance genes in Δ1538 compared to WT after treatment with PMB through qRT-PCR. (d) Heat stress survival assay at (55°C) (e) Expression of SEN1538 in WT after exposure to heat stress through qRT-PCR. (f) Expression of stress response genes in Δ1538 compared to WT after exposure to heat stress through qRT-PCR. (g) Mg2+ starvation stress and survival assay (h) Expression of SEN1538 in WT during Mg2+ starved condition. The bacterial counts were enumerated at indicated time points. Survivals (%) were compared to WT value normalized to 100%. 16 srRNA gene was taken as housekeeping gene in qRT-PCR experiments. Protein levels of 6X-His tag fusion protein of SEN1538 in WT (WT-SEN1538His) strain were assessed during (i) AMP stress (PMB, 1 µg/mL) and (j) heat stress at 55°C by western blot and densitometry analysis. Western blots were scanned by GE Healthcare ImageQuant™ LAS 500 and band intensities were quantified by ImageJTM software. Lane 1, WT-SEN1538His (untreated); lane 2, WT-SEN1538His (stress). Each lane was independently compared to the control after normalization with loading control RpoA. Error bars indicate the mean±SD for three independent experiments. Statistical significance: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant, P ≥ 0.05; Two-way ANOVA (Figure 2(a,d)); Student’s t-test Figure (2(b,c,e,f,h,j))
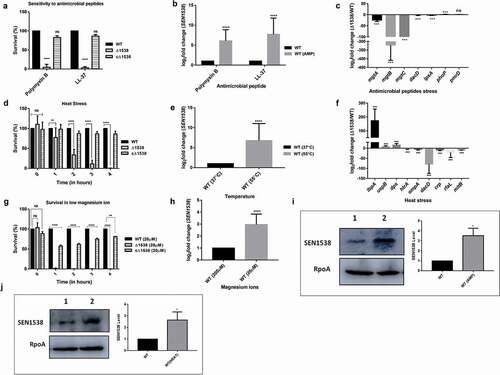
Figure 3. RpoS regulates SEN1538 expression. WT and ΔrpoS were grown at 37°C at 150 rpm until log phase for subsequent experiments. Expression of SEN1538, yciG and ymdF in ΔrpoS compared to WT were checked under different conditions through qRT-PCR (a) LB medium (b) M9 minimal medium (c) Heat stress at 55°C (d) Antimicrobial peptide (AMP) stress (e) Mg2+ starvation stress (f) Bile stress. 16 srRNA gene was taken as housekeeping gene for the analysis. Error bars indicate the mean fold expression ± SD. Statistical significance: **, P < 0.01; ***, P < 0.001; Student’s t-test
Figure 4. Δ1538 displays reduced invasion into HCT116 colon epithelial cells. Adhesion and invasion assay of WT, Δ1538 and the complemented strain cΔ1538 were performed in HCT116 cell line at MOI of 10. (a) Adhesion assays were performed on ice for 30 min. (b) Invasion assays were performed for 50 min. Noninvasive ΔinvC strain served as an experimental control. Data were represented as adhesion (%), invasion (%) for Δ1538, cΔ1538 and were compared to WT value normalized to 100%. (c) Expression of SEN1538 in HCT116 after infection with WT at indicated time points through qRT-PCR. (d) Expression of SPI-1 genes in Δ1538 compared to WT through qRT-PCR. 16s rRNA was taken as housekeeping gene in qRT-PCR analysis. Error bars indicate the mean ± SD of three independent experiments. (e) HCT116 cells were infected with GFP-expressing strains; WT-pCJLA, Δ1538-pCJLA and the complemented strain cΔ1538-pCJLA at MOI of 50. Invasion of strains was measured by the mean fluorescence intensity (MFI) of green fluorescent protein (GFP) expression (%). Data were acquired using BD FACScanto™ II cytometer (Becton–Dickinson, Erembodegem, Belgium) and analyzed by using Flowjo v. 10.4.2. Statistical significance: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant, P ≥ 0.05; One-way ANOVA (Figure 4(a,b)); Student’s t-test (Figure 4(c,d))
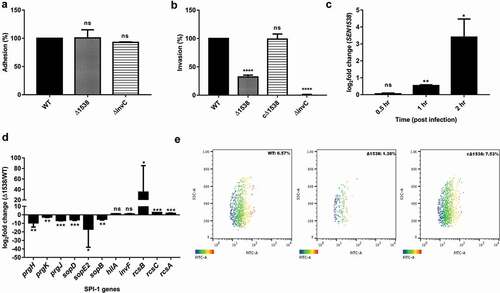
Figure 5. SEN1538 promotes survival within RAW264.7 murine macrophage cells. RAW264.7 cell lines were infected with WT, Δ1538 and the complemented strain cΔ1538 at MOI of 10 for (a) Macrophage uptake assay and (b) Intra-macrophage survival assay. Bacterial counts were enumerated by serial dilution and plating at 2 h and 24 h of infection. Uptake (%) for Δ1538 and cΔ1538 was compared to WT (normalized to 100%). Intra-macrophage survival was represented in fold survival and compared to WT. ΔssaV defective for intra-macrophage survival serves as the experimental control. (c) Expression of SEN1538 in RAW264.7 after infection with WT at indicated time points through qRT-PCR. (d) Expression of SPI-2 genes in Δ1538 compared to WT through qRT-PCR. 16s rRNA was taken as housekeeping gene in qRT-PCR experiments. Error bars indicate the mean ± SD of three independent experiments. (e) Validation of macrophage uptake and survival assay was done by flow cytometry. RAW264.7 cells were infected with GFP-expressing strains; WT-pCJLA, Δ1538-pCJLA and the complemented strain cΔ1538-pCJLA at MOI of 50. Data were acquired at 2 h (uptake) and 24 h (replication survival) using BD FACScanto™ II cytometer (Becton–Dickinson, Erembodegem, Belgium) and analyzed by using Flowjo v. 10.4.2. Statistical significance: *, P < 0.05; **, P < 0.01;***, P < 0.001; ****, P < 0.0001; ns, not significant, P ≥ 0.05; One-way ANOVA (Figure 5(a,b)); Student’s t-test (Figure 5(c,d))
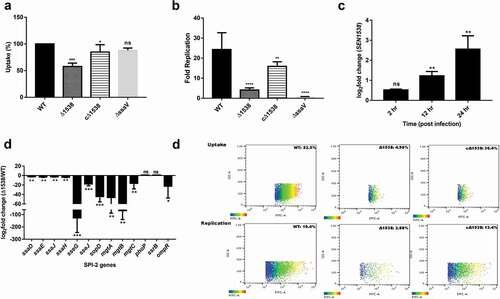
Figure 6. SEN1538 deletion impairs systemic survival and colonization in vivo. Streptomycin-treated C57BL/6 mice (n = 5) were infected with ~107 cfu of WT, Δ1538, the complemented strain cΔ1538 and PBS (negative control) by oral gavage. (a) Bacterial loads in the feces were enumerated at 24 h and at 48 h post-infection (p.i.). (b-e) Organ loads infected with WT, Δ1538 and cΔ1538 were enumerated at 72 h p.i. Mice groups were euthanized and the bacterial load in organs (b) mesenteric lymph node (mLN) (c) spleen (d) liver and (e) cecal content were determined. (f) Lipocalin-2 concentrations from mice feces supernatant were monitored by ELISA. Bar indicates median. (g) Hematoxylin & Eosin (H&E)-stained representative cecum tissue sections (size: 5 µM) from each group of mice showing induced cecal inflammation from WT, Δ1538, and cΔ1538 (left to right). L, lumen; LP, lamina propria; S, submucosal edema. Bars, 200 µM. (h) Cecal pathoscores were obtained by examining the H&E stained cecal tissue sections from each mouse of all groups. Data were represented as mean ± SD. Statistical significance: ns, not significant P ≥ 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; Mann–Whitney U test (Figure 6(a,b,c,d,e,h)); One-way ANOVA (Figure 6(f))
Figure 7. Global changes in the Δ1538 transcriptome compared to WT. (a) Sample replicates of Δ1538 and WT displayed a high degree of correlation with Pearson correlation coefficients ranging from 0.98 to 1. (b) Pie-Donut depicting the top 5 GO and KEGG Pathway illustrating functional clusters that are differentially expressed in Δ1538 against WT. (c) Hierarchical clustering of differentially regulated genes showing sample duplicates of WT and Δ1538. (d-g) Expression profiles of differentially regulated genes in Δ1538 compared to WT, clustered according to their functions. (d) Transporters (e) Pathogenesis (f) Metabolism (g) Hypothetical proteins. Transcriptome data were analyzed and differentially expressed transcripts were selected on the basis of reads per kilobase per million mapped sequence reads (RPKM) >1 in either of the pair of samples with Log2 fold change ≤ −1.5 for downregulated genes and Log2 fold change ≥1.5 for upregulated genes. Statistical significance: t-test P-Value <0.05