Figures & data
Table 1. Primers used in this study
Figure 1. Knockdown of Rab18 inhibits CSFV production. (a–c) Knockdown of Rab18 in SUVECs by lentivirus-mediated shRNA interference. (a) Confirmation of NTshRNA, shRab18-1, shRab18-2, and shRab18-3 cell lines by detection of the enhanced green fluorescent protein (EGFP) reporter. (a) Mock-transfected SUVECs. (b) SUVECs transfected with lentiviruses expressing NTshRNA. SUVECs transfected with lentiviruses expressing (c) shRab18-1, (d) shRab18-2, or (e) shRab18-3. Scale bars, 100 μm. (b) qRT-PCR and western blot analyze Rab18 gene and protein expression levels in Rab18 knockdown cell lines. β-actin served as an internal control. (c) Cell viability of shRab18-3 cell lines. (d) qRT-PCR analyzes CSFV RNA level and TCID50L assay to detect CSFV viral titers in the supernatants of NTshRNA cell lines, shRab18-3 cell lines, and Rab18-mutant-transfected cells. (e) Western blot analyzes Rab18-mutant in transfected cells using an anti-Flag antibody. β-actin served as an internal control
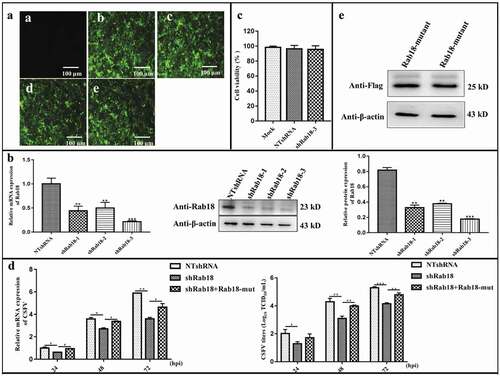
Figure 2. Overexpression of Rab18 enhances CSFV production. (a–c) Overexpression of Rab18 in SUVECs. (a) Confirmation of CMV and CMV-Rab18 cell lines by detection of the enhanced green fluorescent protein (EGFP) reporter. (a) Mock-transfected SUVECs. SUVECs transfected with lentiviruses expressing (b) CMV or (c) CMV-Rab18. Scale bars, 100 μm. (b) qRT-PCR and western blot analyzes Rab18 gene and protein expression levels in Rab18 overexpression cell lines. β-actin served as an internal control. (c) Cell viability of CMV-Rab18 cell lines. (d) qRT-PCR analyzes CSFV RNA level and TCID50 assay to detect CSFV viral titers in the supernatants of CMV cell lines and CMV-Rab18 cell lines. (e) Western blot analyzes pCDNA-Flag-Red-Rab18, pCDNA-Flag-Red-Rab18 (Q67L), and pCDNA-Flag-Red-Rab18 (S22 N) in transfected cells by anti-Flag antibody. β-actin served as an internal control. (f) qRT-PCR analyzes CSFV RNA level and TCID50 assay to detect CSFV viral titers in supernatants of pCDNA-Flag-Red-Rab18-, pCDNA-Flag-Red-Rab18(Q67L)-, and pCDNA-Flag-Red-Rab18(S22N)-transfected SUVECs. β-actin served as an internal control
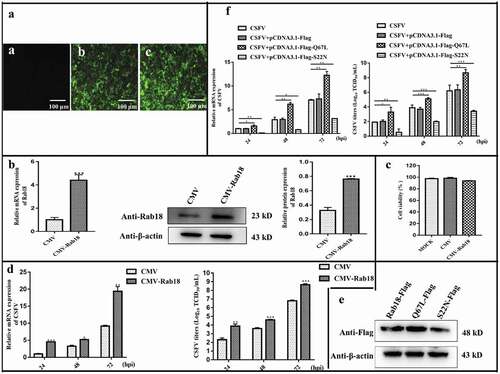
Figure 3. Rab18 is involved in CSFV replication and assembly steps. (a) (a) In the virus binding assay, NTshRNA and shRab18-3 cell lines were incubated with CSFV for 1 h at 4ºC, total cell RNA was extracted, and the CSFV RNA level was quantified by qRT-PCR. β-actin served as an internal control. In the virus entry assay, NTshRNA and shRab18-3 cell lines were incubated with CSFV for 1 h at 4ºC. Then, cells were washed with PBS and cultured in fresh medium at 37ºC for 2 h. Finally, total cell RNA was extracted, and the CSFV RNA level was quantified by qRT-PCR. β-actin served as an internal control. (b) Detection of CSFV entry by IFA. NTshRNA and shRab18-3 cell lines were infected with CSFV (5 MOI) for 1 h at 4ºC. Then, cells were washed with PBS and cultured in fresh medium at 37ºC for 2 h. Finally, cells were fixed with 4% paraformaldehyde, and immunofluorescence staining was performed using anti-E2 antibodies. Cells were counterstained with DAPI to label nuclei (blue). (b) Rab18 involvement in CSFV replication. (a) NTshRNA and shRab18-3 cell lines were incubated with CSFV at different time points (1, 3, 5, 7 h), total cell RNA was extracted, and CSFV RNA level was quantified by qRT-PCR. β-actin served as an internal control. (b) pcDNA-Flag-Red-Rab18-transfected cells were incubated with CSFV (0.5 MOI). At 6 h post-infection, cells were fixed with 4% paraformaldehyde, and immunofluorescence staining was performed using anti-dsRNA antibodies. Cells were counterstained with DAPI to label nuclei (blue). Scale bars, 5 μm. (c) Rab18 involvement in CSFV assembly. (a) NTshRNA and shRab18-3 cell lines were infected with CSFV. At 5 h post-infection, intracellular CSFV RNA levels were determined by qRT-PCR. (b) NTshRNA and shRab18-3 cell lines were infected with CSFV, and 5 h post-infection, cell supernatants were removed and disrupted by multiple cycles of freezing and thawing. The viral titer was determined by the TCID50 assay. (c) Relative intracellular specific infectivity was calculated as a ratio of the intracellular viral infectivity to the intracellular CSFV RNA. (d) NTshRNA and shRab18-3 cell lines were infected with CSFV, and 7 h post-infection, intracellular CSFV RNA levels were determined by qRT-PCR. (e) NTshRNA and shRab18-3 cell lines were infected with CSFV, and 7 h post-infection, cell supernatants were removed and disrupted by multiple cycles of freezing and thawing. The viral titer was determined by the TCID50 assay. (f) Relative intracellular specific infectivity was calculated as a ratio of the intracellular viral infectivity to the intracellular CSFV RNA
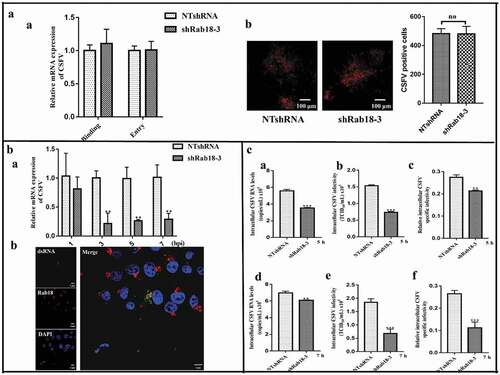
Figure 4. Rab18 binds to CSFV NS5A. (a) Co-immunoprecipitation (co-IP) assay. Cells were transfected with pcDNA-NS2-Myc or pcDNA-NS5A-Myc for 48 h. The transfected cells were lysed and immunoprecipitated, and western blot analysis was conducted using anti-Myc, anti-Rab18, and anti-β-actin. (b) GST-pulldown assay. GST or GST-Rab18 fusion proteins expressed in E. coli BL21 (DE3) were purified with glutathione agarose resin and incubated with the lysate of NS5A-Myc-expressing cells. Western blot analysis using anti-GST, anti-Myc, and anti-β-actin. (c) Exogenous NS5A-Myc binds to Rab18-Flag, S22N-Flag, and Q67 L-Flag in co-transfected cells. pcDNA-NS5A-Myc with pCDNA3.1-Rab18-Flag-, pCDNA3.1-S22N-Flag-, and pCDNA3.1-Q67L-Flag-transfected cells were lysed and immunoprecipitated. Then, western blot analysis was conducted using anti-Myc, anti-Flag, and anti-β-actin. pcDNA-NS2-Myc with pCDNA3.1-Rab18-Flag was used as a negative control. (d) Rab18 co-localization with CSFV NS5A protein. Cells were co-transfected with NS5A-GFP and Rab18-Red, NS5A-GFP and Q67L-Red, and NS5A-GFP and S22N-Red. Plasmids pEGFP-N1 and pCDNA-Red were co-transfected as a control. At 48 h after transfection, cells were fixed in 4% paraformaldehyde and stained with DAPI to label nuclei (blue). Scale bars, 10 μm
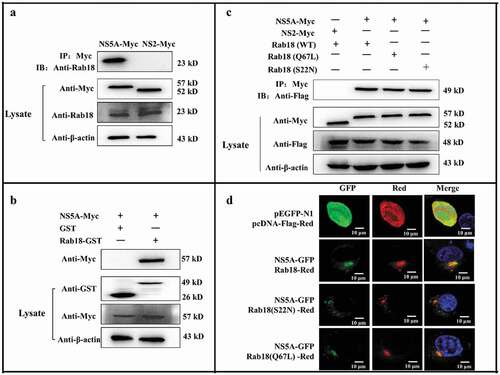
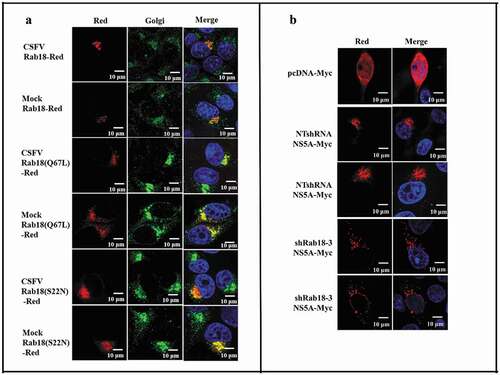
Figure 5. Redistribution of NS5A in Rab18 knockdown cells. (a) Rab18 localization at the Golgi apparatus. Cells seeded on glass coverslips were transfected with a Rab18-Red, Rab18 (Q67L)-Red, or Rab18 (S22N)-Red expression plasmid, as indicated. At 48 h after transfection, cells were fixed in 4% paraformaldehyde, and immunofluorescence staining was performed using an anti-TGN46 antibody. Cells were also counterstained with DAPI to label nuclei (blue). Scale bars, 10 μm. (b) NS5A distribution in a punctate pattern. NTshRNA and shRab18 cell lines seeded on glass coverslips were respectively transfected with pcDNA-Myc and NS5A-Myc expression plasmids, as indicated. At 48 h after transfection, cells were fixed in 4% paraformaldehyde, and immunofluorescence staining was performed using an anti-Myc antibody. Cells were also counterstained with DAPI to label nuclei (blue). Scale bars, 10 μm