Figures & data
Table 1. Primers and probes*.
Figure 1. Preparation of plasmid standards. A-D: amplification curves (X-axis: Cycle, Y-axis: Fluorescence) of PEDV, PDCoV, PToV, and SADS-CoV for each plasmid standard of concentrations with 1 × 107 copies/μL to 1 × 101 copies/μL; E-H: standard curves of plasmid standards of PEDV, PDCoV, PToV, and SADS-CoV. All standard curves were conducted with software GraphPad Prism 8.
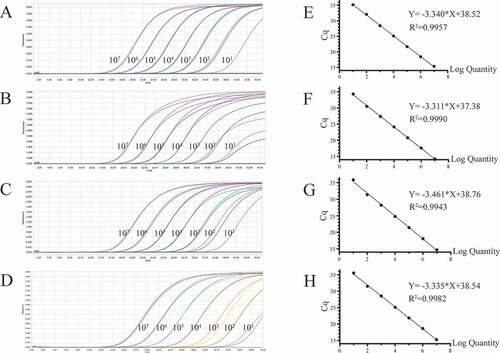
Table 2. Cq values of PEDV, PDCoV, PToV, and SADS-CoV detected by this multiplex real-time PCR assay with different probe and primer concentrations.
Figure 2. A-D: amplification curves (X-axis: Cycle, Y-axis: Fluorescence) of PEDV, PDCoV, PToV, and SADS-CoV detected by multiplex real-time PCR with different probe and primer concentrations. Plasmid standards with concentration of 1 × 103 copies/μL were chosen as templates for the reactions. The four green lines are the amplification curves of four fluorescence of the most suitable reaction tube.
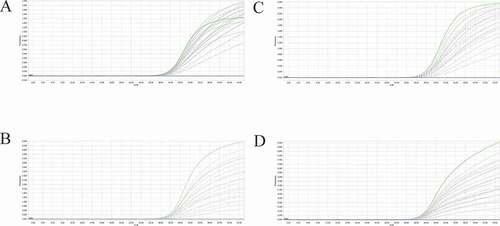
Table 3. Sensitivity of the multiplex real-time PCR assay.
Figure 3. A: amplification curves (X-axis: Cycle, Y-axis: Fluorescence) of 10-fold serial dilutions (1 × 107–1 × 101 copies/μL) of plasmid standards of PEDV, PDCoV, PToV, and SADS-CoV detected by multiplex real-time PCR. B: four amplification curves represent samples positive for PEDV, PDCoV, PToV, and SADS-CoV detected by our multiplex real-time PCR assay; negative samples include TGEV, PoRV, PSV, PTV, CSFV, PKV, and negative control.
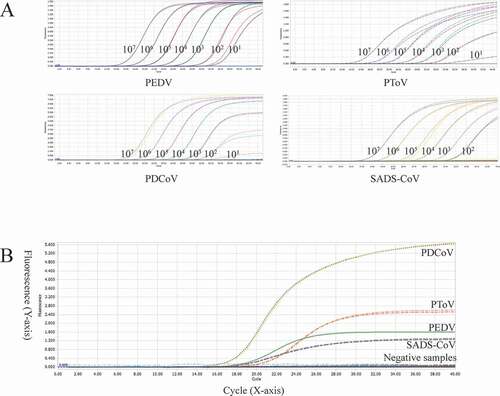
Figure 4. Co-infection simulation experiments with two pathogens. A-F: amplification curves (X-axis: Cycle, Y-axis: Fluorescence) of PDCoV + SADS-CoV, PDCoV + PToV, PEDV + SADS-CoV, PEDV + PDCoV, PEDV + PToV, PToV + SADS-CoV at concentrations of 1 × 103 copies/μL; G-L: amplification curves of PDCoV + SADS-CoV, PDCoV + PToV, PEDV + SADS-CoV, PEDV + PDCoV, PEDV + PToV, PToV + SADS-CoV at concentrations of 1 × 102 copies/μL. Two replicates were set per reaction.
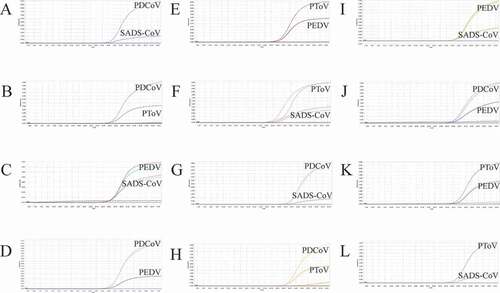
Figure 5. Co-infection simulation experiments with three pathogens. A-D: amplification curves (X-axis: Cycle, Y-axis: Fluorescence) of PDCoV + PToV + SADS-CoV, PEDV + PToV + SADS-CoV, PEDV + PDCoV + SADS-CoV, PEDV + PDCoV + PToV with concentration of 1 × 103 copies/μL; E-H: amplification curves of PDCoV + PToV + SADS-CoV, PEDV + PToV + SADS-CoV, PEDV + PDCoV + SADS-CoV, PEDV + PDCoV + PToV with concentrations of 1 × 102 copies/μL. Two replicates were set per reaction.
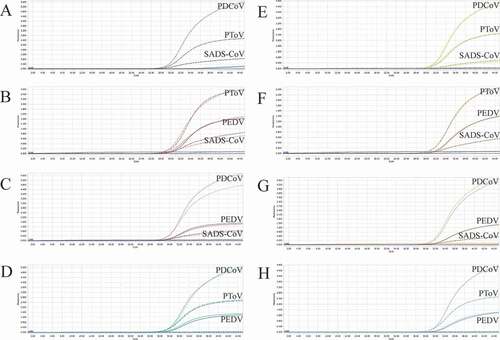
Figure 6. Co-infection simulation experiments with four pathogens. A-B: amplification curves (X-axis: Cycle, Y-axis: Fluorescence) of PEDV + PDCoV + PToV + SADS-CoV at concentrations of 1 × 102 copies/μL and 1 × 107 copies/μL. Two replicates were set per reaction.
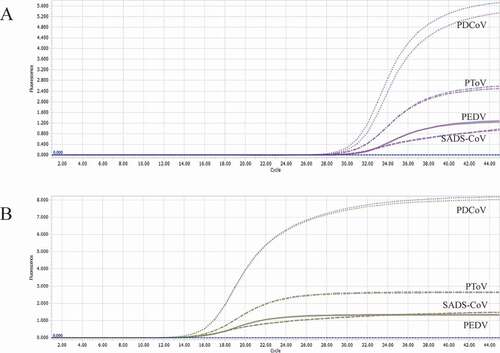
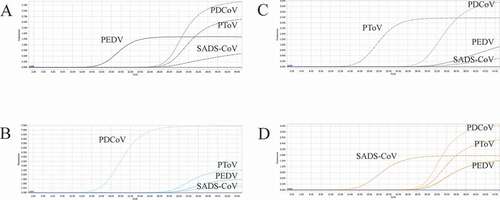
Figure 7. Co-infection of all the four pathogens at different concentrations. A: the concentration of plasmid standard of PEDV was 1 × 107 copies/μL and the others were 1 × 102 copies/μL; B: the concentration of plasmid standard of PDCoV was 1 × 107 copies/μL and the others were 1 × 102 copies/μL; C: the concentration of plasmid standard of PToV was 1 × 107 copies/μL and the others were 1 × 102 copies/μL; D: the concentration of plasmid standard of SADS-CoV was 1 × 107 copies/μL and the others were 1 × 102 copies/μL. Two replicates were set per reaction. X-axis: Cycle, Y-axis: Fluorescence.