Figures & data
Table 1. Oligonucleotides used in this study.
Figure 1. Sequence analysis of the Cu transporter CrpF. Proposed two-dimensional model of CrpF (FOXG_03265; XP_ 018237293.1) protein describing predicted metal-binding motifs (metal-binding domains or heavy metal associated domains), conserved functional domains (phosphatase domain, Cu translocation domain, aspartyl kinase domain, ATP binding domain) and structural domains (8 transmembrane domains) and comparison of the conserved functional domains among different species. GenBank accession numbers are given in parentheses: A. fumigatus CrpA (AFUA_3G12740; XP_754347.1), Cryptococcus neoformans (OXG38219.1), Neurospora crassa (XP_957691.3) and Mucor circinelloides (OAD01460.1).
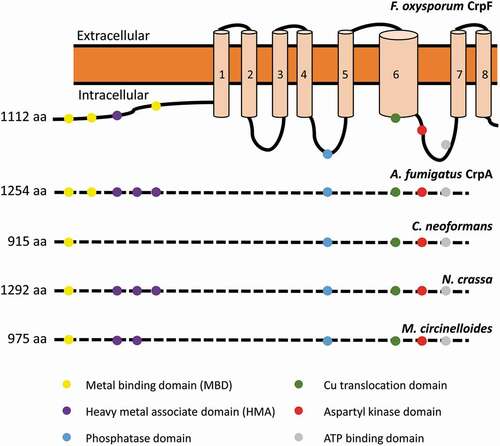
Figure 2. F. oxysporum contains a putative Cu-exporting ATPase in its genome. (a) Alignment of the amino acid sequences of the predicted Cu-exporting ATPases encoded by gene FOXG_ 03265 (CrpF) and the corresponding orthologous gene crpA from A. fumigatus using Clustal method. Protein accession numbers are reported as follows: A. fumigatus CrpA (AFUA_3G12740) and F. oxysporum CrpF (FOXG_03265). (b) Phylogenetic tree of predicted Heavy-metal ATPases (HMA) in fungal species distributed in clades Cc2-type, Crp-type and Pca-type. The phylogenetic tree depicts clades of HMA protein sequences available at NCBI (sequences ID are shown in parenthesis). HMA proteins are highly conserved among fungi kingdom. In green are highlighted the F. oxysporum proteins. Human orthologous gene AAA35580.1 is also included. Phylogram was constructed using Neighbour-Joining method. Bootstrap values obtained from 1000 replicates are indicated at the nodes. Scale bar indicates the relative length of each branch. Clustal W was used for protein alignment.
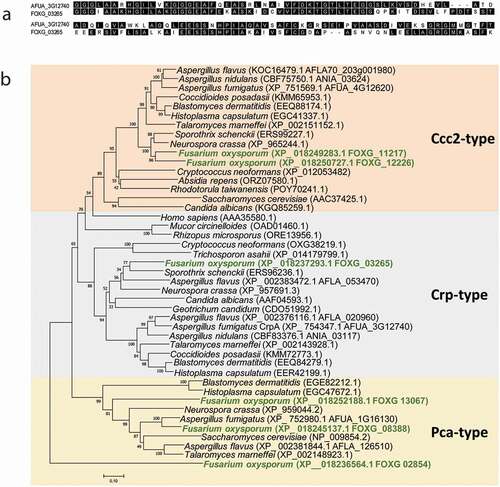
Figure 3. Targeted deletion of the F. oxysporum crpF gene. (a) Targeted gene replacement strategy using a disruption construct obtained by fusion PCR with HygR cassette as selective marker. Relative positions of primers used for PCR and probe (dashes bars) are indicated. (b) Southern blot analysis of gDNA from wt strain and transformants. gDNAs were digested with Sma I and Sac II to detect deletion and complementation of crpF. The Southern blot image provided comes from two nitrocellulose membranes. The images were manipulated with the objective of only show the interesting transformants.
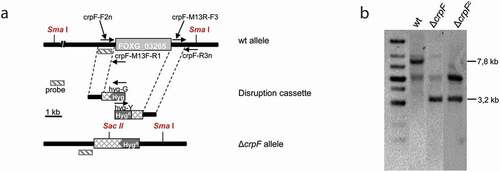
Figure 4. Loss of crpF impairs growth of F. oxysporum under Cd, Cu and Zn exposure. Fungal colonies from wt, ΔcrpF and complemented ΔcrpFc strains grown for 6–7 days at 28°C on SM plates containing 0.15 mM Cd (CdCl2), 0.4 mM Cu (CuSO4) or 20 mM Zn (ZnCl2). The number of inoculated spores is indicated.
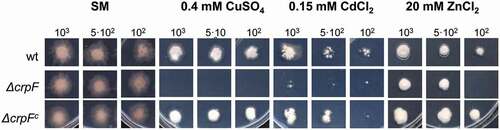
Figure 5. Intracellular Cu determination. Cu concentration was quantified on mycelia from wt and ΔcrpF strains grown in SM (control) and SM supplemented with 0.1 mM CuSO4. The amount of Cu is reported as mg g−1 biomass. Values are the average of three replicates.
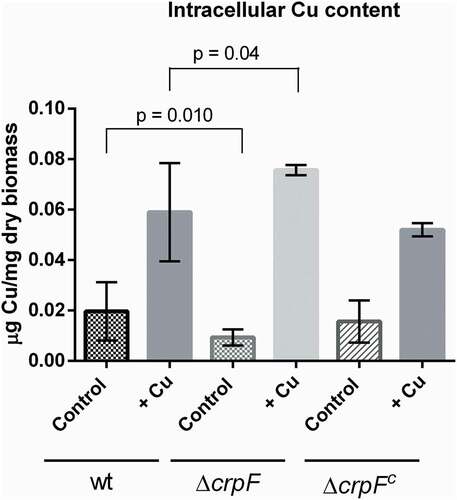
Figure 6. Transcriptional analyses of crpF, metal homeostasis mt1 and aceA and stress prx and gapdh related genes by RT-PCR analysis. Transcript levels of act, mt1, crpF, aceA, prx and gapdh from wt and ΔcrpF strains on control condition and under exposure to 0.1 mM CdCl2, 0.175 mM CuSO4 or 7.5 mM ZnCl2 are shown.
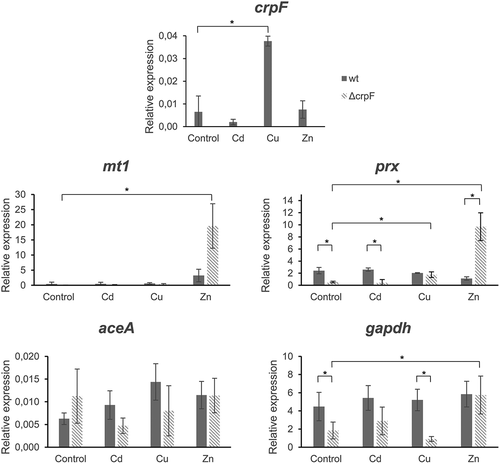
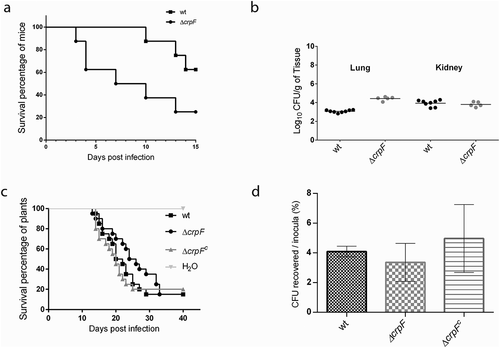
Figure 7. Pathogenic behaviour of ΔcrpF in different infection models. (a) Virulence of F. oxysporum strains in immunosuppressed murine model host inoculated with 107 microconidia of the indicated strains by lateral tail vein injection. Percentage survival values were plotted for 15 days. (b) Fungal load quantification of lung and kidney from eight randomly chosen surviving mice from each strain at day 7 after infection using CFU estimation method. (c) Gene crpF is not required for virulence of F. oxysporum in a plant model of infection. Survival of groups of 10 tomato plants inoculated by immersing the roots into a suspension of 5 × 106 freshly obtained microconidia mL−1 of the indicated strains. Experiments were performed at least three times with similar results and the percentage survival values were plotted for 30 days from one representative experiment. (d) Percentage of viable hypha recovered after 6 h of co-cultivation with macrophages. Data are means of results from duplicated samples from three independent experiments.