Figures & data
Figure 1. Organization of the MGI of N. meningitidis strain B16B6. Strain B16B6 has three MGI [Citation10]. Genes flanking the islands are indicated with filled black arrows. Filled colored arrows indicate mafA and mafB genes; their proposed systematic names [Citation11] and the phylogenetic classes of the encoded proteins [Citation10] are given above and below the arrows, respectively. Color coding: orange, MafAI; blue, MafAII; garnet, MafBI; pink, MafBII; green, MafBIII. Open colored arrows downstream of the mafB genes refer to truncated mafB genes that lack regular 5ʹ sequences. Their color refers to high sequence similarity with the upstream intact mafB, although their 3ʹ end, encoding the toxic domain, is entirely different. MGI-3 contains an additional complete mafB gene, but it is disrupted by a frameshift mutation (indicated with red slash). Open black arrows represent small open reading frames, including mafI genes encoding immunity proteins. Where multiple small open reading frames are located downstream of a mafB or truncated mafB, it is not always clear which one encodes the corresponding immunity protein. Lines numbered a-d refer to the polypeptides that were produced to raise antisera
![Figure 1. Organization of the MGI of N. meningitidis strain B16B6. Strain B16B6 has three MGI [Citation10]. Genes flanking the islands are indicated with filled black arrows. Filled colored arrows indicate mafA and mafB genes; their proposed systematic names [Citation11] and the phylogenetic classes of the encoded proteins [Citation10] are given above and below the arrows, respectively. Color coding: orange, MafAI; blue, MafAII; garnet, MafBI; pink, MafBII; green, MafBIII. Open colored arrows downstream of the mafB genes refer to truncated mafB genes that lack regular 5ʹ sequences. Their color refers to high sequence similarity with the upstream intact mafB, although their 3ʹ end, encoding the toxic domain, is entirely different. MGI-3 contains an additional complete mafB gene, but it is disrupted by a frameshift mutation (indicated with red slash). Open black arrows represent small open reading frames, including mafI genes encoding immunity proteins. Where multiple small open reading frames are located downstream of a mafB or truncated mafB, it is not always clear which one encodes the corresponding immunity protein. Lines numbered a-d refer to the polypeptides that were produced to raise antisera](/cms/asset/24a273d8-1658-44f9-8df6-c5531ca9e16f/kvir_a_1851940_f0001_oc.jpg)
Table 1. Bacterial strains and plasmids used in this study
Figure 2. Expression, secretion, and proteolysis of MafBI*. (a) Structure of MafBI.1 encoded by locus NMC1790 of strain FAM18. The protein contains a signal sequence (SS), a DUF1020 domain, a Hint domain, and a C-terminal toxic domain. The mass of each domain is given (in kDa). The recombinant MafBI* proteins lack the toxic domain and instead contain either a Strep-tag or, where explicitly indicated, a His-tag. (b) Secretion and processing of MafBI*. Proteins in cell lysates (c) and spent medium (m) of BB-1 (WT) and its single and double mafAI mutant derivatives (ΔAI.1 and ΔAI.2) were separated by SDS-PAGE and probed on Western blots with antisera directed against MafB (α-MafBI) or against the His-tag (α-His). The presence or absence in the strains of pEN-mafBI-S encoding MafBI* with a Strep-tag in the left panel or of pEN-mafBI-H, which encodes a His-tagged variant of MafBI*, in the right panel and of IPTG to induce MafBI* production, is indicated. Open arrow-heads indicate MafB forms suggested to be the precursor (p), the mature form after removal of the signal sequence (M), the DUF1020 domain (d), and proteolytic DUF1020 fragments (f). The filled arrow-head indicates a cross-reacting band. (c) The medium fractions (M) of the wild-type and the mafAI double mutant, both expressing MafBI*, were subjected to ultracentrifugation, and the pelleted OMVs (o) and the OMV-depleted supernatant (M−) were analyzed by SDS-PAGE and Western blotting. As controls, the presence in the fractions of the OM-associated periplasmic protein RmpM and the secreted α-peptide (αP) of IgA protease were analyzed. (d) Protease accessibility of MafBI* forms. Upper panels: Intact cells of the parent strain (WT) and the ΔmafAI double mutant, both synthesizing MafBI* from pEN-mafBI-S, were incubated with 2 µg ml−1 of proteinase K. Lower panels: spent-medium fractions of the same cultures were treated with proteinase K. Degradation of various proteins was assessed by Western blotting. Different MafBI* forms are indicated at the right and labeled as in panel B. (e) The medium fraction of the ΔmafAI double mutant producing MafBI* was either treated or not with 1% elugent before proteinase K digestion
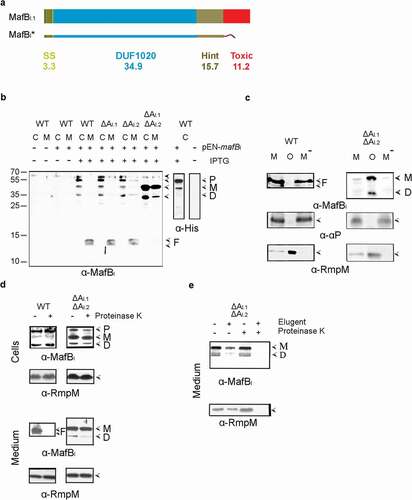
Figure 3. Expression, secretion, and proteolysis of MafBIII. (a) Structure of MafBIII encoded on MGI-3 of strain B16B6. The protein contains a signal sequence (SS), a DUF1020 domain, and a C-terminal toxic domain. The mass of each domain is given (in kDa). The truncated MafBIII* protein lacks the toxic domain and instead contains a Strep-tag. (b) Expression and localization of MafBIII* in N. meningitidis. Proteins in cell lysates (c) and spent medium (m) of the unencapsulated strain BB-1 (WTc-), its derivative lacking mafAII (ΔAII), and a derivative of the encapsulated parental strain B16B6 (c+) lacking all mafA genes (ΔAI.1ΔAI.2ΔAII) were separated by SDS-PAGE and probed on Western blots with antiserum directed against MafBIII. The presence of plasmid pEN-mafBIII-S in the strains and of IPTG during growth is indicated. (c) The medium fraction (M) of strain BB-1 expressing MafBIII* was subjected to ultracentrifugation, and the pellet containing OMVs (o) and the OMV-depleted supernatant (M−) were analyzed by SDS-PAGE and Western blotting. As controls, the presence in the fractions of the OM-associated protein RmpM and the secreted α-peptide (αP) of IgA protease was analyzed. (d) Culture medium from strain BB-1 expressing MafBIII* was treated with 2 µg ml−1 proteinase K, and degradation of MafBIII* and, as a control, of the OM-associated periplasmic protein RmpM, was assessed by Western blotting. In panels b-d, F indicates proteolytic fragments of MafBIII.
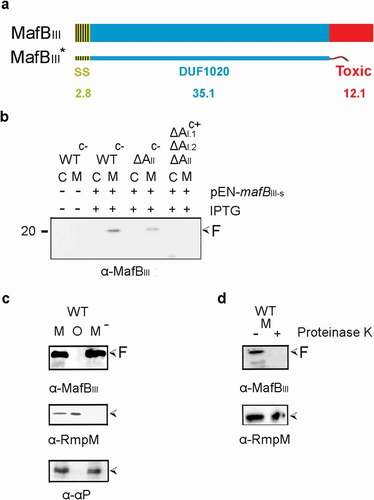
Figure 4. Expression and localization of MafBIII in E. coli. (a) MafBIII and the corresponding MafI were produced in BL21(DE3) either with (BL21-AII+BIII+I) or without (BL21-BIII+I) MafAII, and cell lysates were analyzed by Western blotting with antiserum against MafBIII. The blot at the right was exposed longer than the one at the left to obtain visible MafB-specific signals. (b) Intact cells of both BL21(DE3) derivatives were incubated with the indicated concentrations of proteinase K and degradation of MafBIII, and, as a control, of OmpA, a β-barrel OM protein with a large periplasmic domain, was assessed by Western blotting. In both panels, M indicates the full-length mature form of MafBIII, and F in panel A indicates proteolytic fragments of the protein. The filled arrow-head in panel A indicates a background band
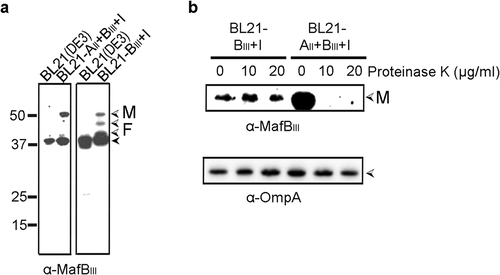
Figure 5. Synthesis and subcellular localization of MafA. (a) The synthesis and possible secretion of MafA proteins were examined by analyzing cell lysates (c) and spent media (m) of BB-1 (WT) and various ΔmafA mutant derivatives on Western blots with anti-MafA antisera. The positions of MafA proteins are indicated with open arrowheads. A faint cross-reactive band with a similar electrophoretic mobility is visible in the mafAI.1 mutant and the double mutant and is indicated with a black arrowhead. (b) Membranes of BB-1 synthesizing a His-tagged recombinant MafAI protein from plasmid pEN-mafAI-H were separated by sucrose density gradient centrifugation, and the fractions obtained were analyzed by Western blotting with antibodies directed against the His-tag, against RmpM as an OM marker, and against DsbA as IM markers. (c) Intact cells or cell envelopes of BB-1ΔnalP expressing His-tagged MafAI were treated with 2 µg ml−1 proteinase K as indicated. Cell envelopes were treated or not with 1% elugent before proteinase K digestion. After incubation, the degradation of MafAI, RmpM and the α-peptide (αP) of IgA protease were assessed by Western blotting using specific antibodies. The αP is exposed at the cell surface covalently connected to the translocator domain (TD) of IgA protease [Citation22]. For this blot, an antiserum directed against the TD was used. (d) Western blot analysis of cell envelopes from strain BB-1 synthesizing or not His-tagged MafAI from plasmid pEN-mafAI-H and MafAII from the chromosome. The blots were probed with anti-His-tag antibodies and anti-MafAII antiserum, respectively. Before electrophoresis, proteins were either denatured or not by heating the samples at 95°C. HMW complex indicates the position of the high-molecular-weight complexes of the MafA proteins detected in the unheated samples. Filled arrowheads mark weak cross-reactive bands. Numbers at the left indicate the positions of molecular mass markers when required. In panels b and c, only relevant parts of the blots are shown
![Figure 5. Synthesis and subcellular localization of MafA. (a) The synthesis and possible secretion of MafA proteins were examined by analyzing cell lysates (c) and spent media (m) of BB-1 (WT) and various ΔmafA mutant derivatives on Western blots with anti-MafA antisera. The positions of MafA proteins are indicated with open arrowheads. A faint cross-reactive band with a similar electrophoretic mobility is visible in the mafAI.1 mutant and the double mutant and is indicated with a black arrowhead. (b) Membranes of BB-1 synthesizing a His-tagged recombinant MafAI protein from plasmid pEN-mafAI-H were separated by sucrose density gradient centrifugation, and the fractions obtained were analyzed by Western blotting with antibodies directed against the His-tag, against RmpM as an OM marker, and against DsbA as IM markers. (c) Intact cells or cell envelopes of BB-1ΔnalP expressing His-tagged MafAI were treated with 2 µg ml−1 proteinase K as indicated. Cell envelopes were treated or not with 1% elugent before proteinase K digestion. After incubation, the degradation of MafAI, RmpM and the α-peptide (αP) of IgA protease were assessed by Western blotting using specific antibodies. The αP is exposed at the cell surface covalently connected to the translocator domain (TD) of IgA protease [Citation22]. For this blot, an antiserum directed against the TD was used. (d) Western blot analysis of cell envelopes from strain BB-1 synthesizing or not His-tagged MafAI from plasmid pEN-mafAI-H and MafAII from the chromosome. The blots were probed with anti-His-tag antibodies and anti-MafAII antiserum, respectively. Before electrophoresis, proteins were either denatured or not by heating the samples at 95°C. HMW complex indicates the position of the high-molecular-weight complexes of the MafA proteins detected in the unheated samples. Filled arrowheads mark weak cross-reactive bands. Numbers at the left indicate the positions of molecular mass markers when required. In panels b and c, only relevant parts of the blots are shown](/cms/asset/9485f9ec-86ec-45cc-93d8-674241fc9b5f/kvir_a_1851940_f0005_b.gif)
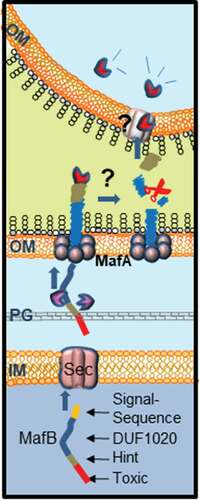
Figure 6. Proposed model for MafB secretion and activation. The MafB proteins contain a cleavable N-terminal signal sequence (yellow) for translocation across the inner membrane (IM) via the Sec translocon. After periplasmic transit, possibly escorted by chaperones like SurA or Skp (purple), MafB interacts with the OM-integrated MafA protein, presumably through its DUF1020 domain. The oligomeric MafA protein constitutes the channel in the OM through which MafB is translocated. After secretion, MafB presumably remains associated with the OM until it interacts with a receptor at the target cell. Then, MafB is cleaved by an unknown protease or via an autocatalytic process that is induced by conformational changes in the protein upon receptor interaction. The C-terminal domain of MafB is thereby released and imported into the target cell where it exerts its toxic activity