Figures & data
Figure 1. Fiber-2 efficiently interacted with host KPNA3/4. (a-b) LMH cells infected with FAdV-4 were immunoprecipitated with mAb 3C2 against Fiber-2. The cell lysates and the immunoprecipitates were examined with western blot using polyclonal antibodies against KPNA3 (a) and KPNA4 (b). (c-d) LMH cells transfected with pcDNA3.1-F1 or pcDNA3.1-F2 were immunoprecipitated with polyclonal antibodies against KPNA3 (c) and KPNA4 (d). The cell lysates and the immunoprecipitates were examined with western blot using chicken sera against FAdV-4. All experiments were performed for three times with comparable results
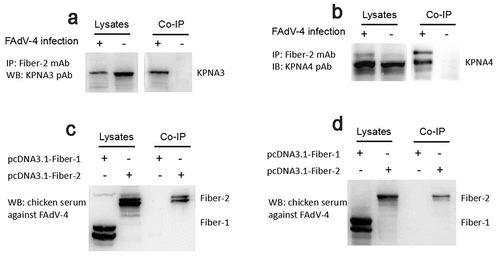
Figure 2. KPNA3/4 assisted viral replication of FAdV-4 in LMH cells. (a-b) Overexpression of KPNA3 and KPNA4 increased viral replication in LMH cells. (a) LMH cells were first transfected with pcDNA3.1-KPNA3, pcDNA3.1-KPNA4 and pcDNA3.1, respectively. 24 hours after transfection, the LMH cells were infected with FAdV-4, and the supernatants collected from infected LMH cells at indicated time points were then titrated with TCID50. The overexpression of KPNA3 and KPNA4 in LMH cells were examined with western blot by polyclonal antibodies against KPNA3 (b-a) and KPNA4 (b-b). (c-d) Knockout of KPNA3 or KPNA4 inhibited viral replication in LMH cells. (c) KPNA3-KO LMH cells, KPNA4-KO LMH cells and LMH cells were infected with FAdV-4, and the supernatants collected from infected LMH cells at indicated time points were then titrated with TCID50. The expression of KPNA3 and KPNA4 in KPNA3-KO and KPNA4-KO LMH cells were examined with western blot by polyclonal antibodies against KPNA3 (d-a) and KPNA4 (d-b), respectively. All experiments were done in triplicates and repeated twice
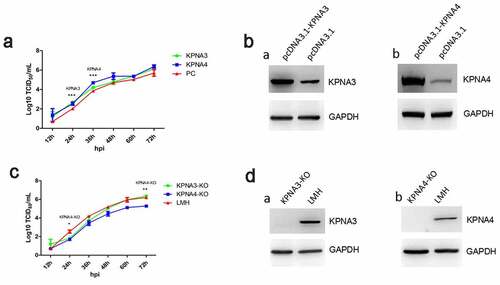
Figure 3. N-terminal 1–40aa in Fiber 2 responsed for the interaction with KPNA3/4. LMH cells were transfected with full-length fiber-2, truncated fiber-2 (1–40 aa deletion), truncated fiber-2 (61–114 aa deletion) and pcDNA3.1, respectively. (a) Cell lysates were prepared and immunoprecipitated with mAb 3C2 against Fiber-2, the pellets were examined with western blot by polyclonal antibodies against KPNA3 and KPNA4. (b) Sequence alignment analysis for N-terminus of 1–40aa in Fiber-2 from different FAdV-4 isolates. All experiments were performed for three times with comparable results
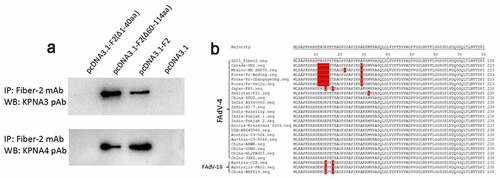
Figure 4. Generation of FAV4_Del carrying Fiber-2 without N-terminal 7–40aa. (a) Strategy of the CRISPR/Cas9 platform for generating the fiber-2-edited virus FAV4_Del. LMH cells were first transfected with sgRNA1 and sgRNA2, and then LMH cells were infected with FAdV-4-EGFP and transfected with donor plasmid at 24 hours post-transfection. The fiber-2-edited virus FAV4_Del was then purified by limiting dilution assay and viral plaque assay. (b) PCR identification of the fiber-2-edited virus FAV4_Del. Viral genome of purified FAV4_Del, FAdV-4 and unpurified FAV4_Del were extracted and identified by PCR. (c) Western blot analysis of the fiber-2-edited virus FAV4_Del. LMH cells were infected with the fiber-2-edited virus FAV4_Del and FAdV-4. 4 d post-infection, the infected LMH cells were harvested and lysed, and the lysates were then examined with western blot by mAb 1C9 against Fiber-2. Experiments B and C were performed for three times with comparable results
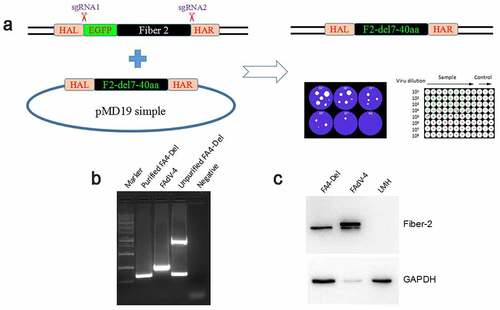
Figure 5. Evaluation of the interaction between KPNA3/4 and FAV4-Del. LMH cells were infected with FAdV-4 and FAV4-Del at 0.1 MOI respectively, and lysed and immunoprecipitated with KPNA3 (a) or KPNA4 (b) polyclonal antibodies, and then the cell lysates were detected with mAb 1C9 against Fiber-2, while the immunoprecipitates were detected with polyclonal antibodies against KPNA3(A) and KPNA4 (B), and mAb 1C9 against Fiber-2, respectively
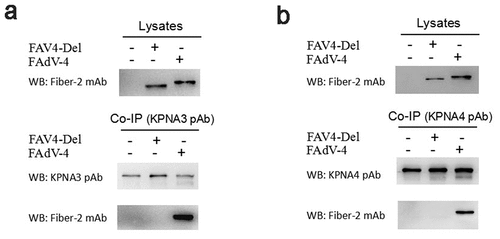
Figure 6. FAV4_Del showed low replication ability in LMH cells. (a) LMH cells were infected with FAV4_Del and FAdV-4 at the same dose, the viral supernatant collected from infected LMH cells at indicated time points were then titrated with TCID50. (b) LMH cells infected with the FAV4_Del and FAdV-4 were analyzed by IFA using mAb 3B5 against Fiber-1. (c) LMH cells infected with FAV4_Del and FAdV-4 were harvested at different time points and lysed by lysis buffer, and then the lysates were examined with western blot by mAb 1C9 against Fiber-2. All experiments were performed for three times with comparable results
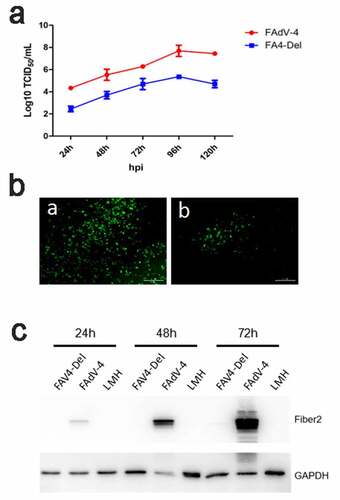
Figure 7. FAV4_Del was highly attenuated in vivo. All the SPF chickens were randomly divided into three groups, and inoculated with FAV4_Del, wild type FAdV-4 and 1% culture medium (NC) respectively. All the SPF chickens were observed for 14 d. (a) Percent of survival for the infected chickens. (b) Representative histological changes in liver tissues from chickens infected with FAdV-4 (a) and FAV4_Del (b). (c) Viral shedding in cloacal swabs from the infected chickens. Viral loads in liver (d), spleen (e) and kidney (f) tissues from the infected chickens
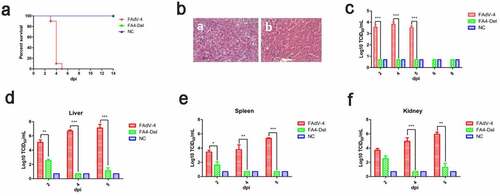
Figure 8. FAV4_Del provided efficient protection against lethal challenge. The chickens inoculated with FAV4_Del and 1% culture medium survived after 21 d were then challenged with the lethal dose of FAdV-4. (a) Percent of survival for the challenged chickens. (b) Representative gross lesion in heart and liver from the challenged control chickens (a) and the challenged chickens previously inoculated with FAV4_Del (b). (c) Viral shedding in cloacal swabs from the challenged chickens. Viral loads in liver (d), spleen (e) and kidney (f) tissues from the challenged chickens
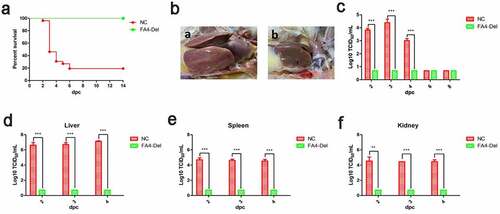
Table 1. PCR primers for plasmids construction
Table 2. Primers used for sgRNA cloning
Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
