Figures & data
Figure 1. Recombinant arginine-rich C-terminal domain of the PE_PGRS3 promotes Mycobacterium smegmatis adhesion to macrophages and pneumocytes
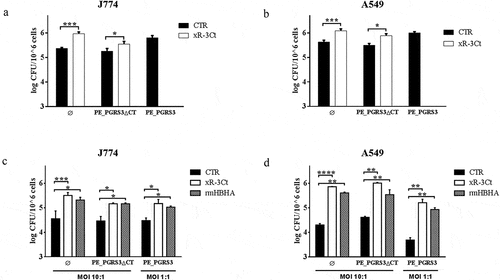
Figure 2. Arginine-rich C-terminal domain specifically binds cardiolipin and phosphorylated phosphatidylinositols
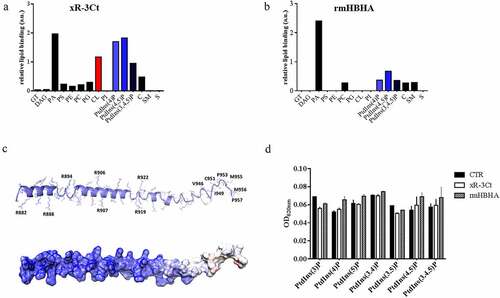
Figure 3. Administration of cardiolipin, but not phosphorylated phosphatidylinositols, to phosphate-starved culture turns off pe_pgrs3 expression
Figure 4. Over-expression of the PE_PGRS3 enhances Mycobacterium tuberculosis cell entry in pneumocytes, but not in murine macrophages or human peripheral blood mononuclear cells (PBMCs)
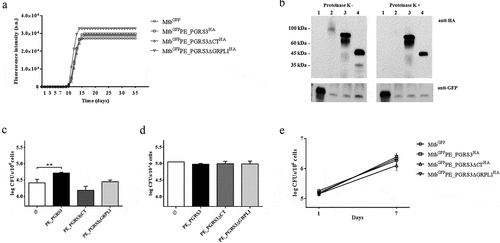
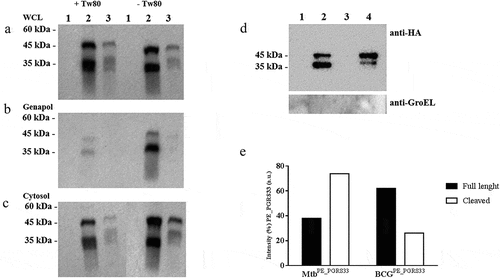
Figure 5. PE_PGRS3 is cleaved in Mycobacterium tuberculosis.
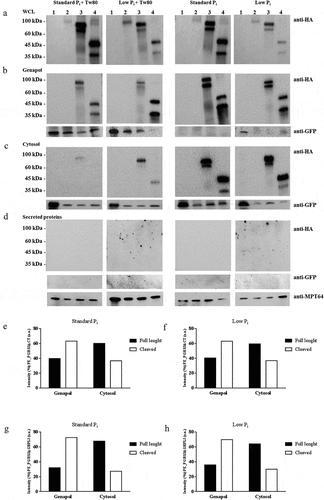
Figure 6. PE_PGRS33 is mainly cleaved in Mycobacterium tuberculosis compared to Mycobacterium bovis BCG
Supplemental Material
Download Zip (2.4 MB)Data Availability Statement:
Raw data were generated at Università Cattolica del Sacro Cuore and Fondazione Policlinico Gemelli (Dipartimento di Scienze biotecnologiche di base, cliniche intensivologiche e perioperatorie – Sezione di Microbiologia; Dipartimento di Diagnostica per Immagini, Radioterapia Oncologica ed Ematologia; Dipartimento di Neuroscienze, Università Cattolica del Sacro Cuore) and Institute of Biostructures and Bioimaging. Derived data supporting the findings of this study are available from the corresponding author GD on request.
