Figures & data
Figure 1. Identification of the region of Tip critical in PilB inhibition
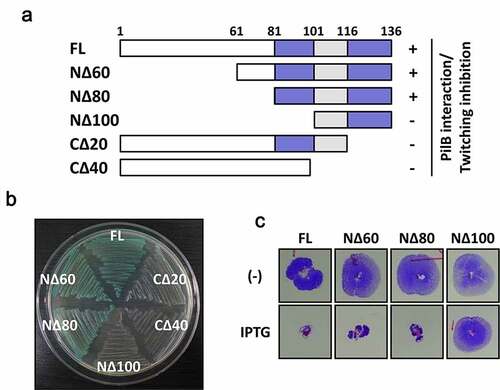
Figure 2. Design and synthesis of a series of Tip-derived peptides
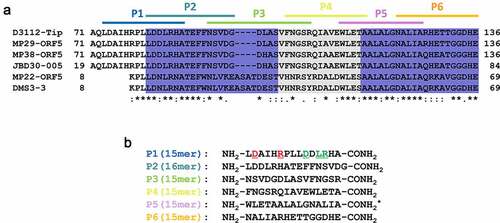
Figure 3. Bioactivity of the Tip-derived peptides upon exogenous administration
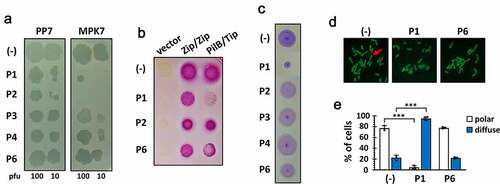
Figure 4. Structure and membrane permeability of P1
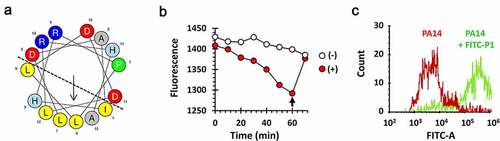
Figure 5. Amino acid residues in P1 critical for the Tip function
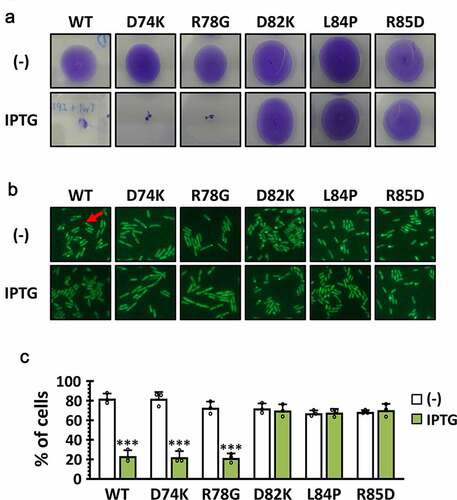
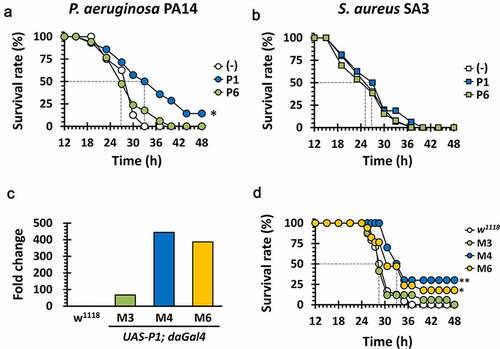
Figure 6. Antibacterial efficacy of P1 in vivo