Figures & data
Table 1. Wild-type and mutant strains of U. virens used in this study
Figure 1. Identification of UvCGBP1 in U. virens. (a) NJ tree of putative CGBP1 homologues from 13 fungal genomes generated with MEGA7.0. The bootstrap percentage values from 1000 repeats are shown at branch nodes. (b) Predicted Pfam domain of C2H2 zinc finger (box) protein UvCGBP1. (c) Transactivation analysis of UvCGBP1 in yeast. The vectors pGBKT7-53/pGADT7-T and pGBKT7 were expressed in yeast as positive and negative controls, respectively. (d) Subcellular localization of UvCGBP1 in U. virens. DIC, differential interference contrast; GFP, green fluorescent protein. Scale bar = 10 μm. (e) qRT-PCR detected expression of UvCGBP1 in conidia (Co), hyphae (Hy) and different infection stages on rice spikelets (1–20 d). (f) Expression analysis of UvCGBP1 after treated with rice leaf and spikelet extracts with qRT-PCR. (g) Expression analysis of UvCGBP1 after treated with chemical, 1 M NaCl or 10 mM H2O2 with qRT-PCR. Asterisks represent significant differences between PSB control and stress treatments based on LSD at P = 0.05
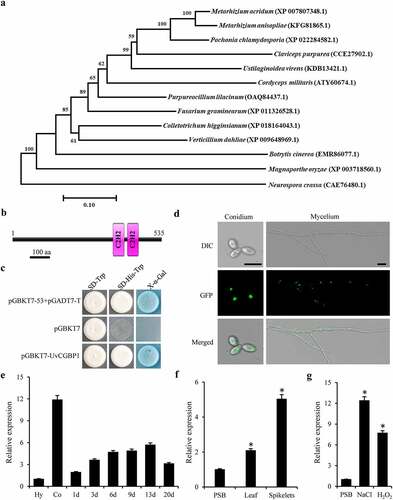
Figure 2. Deletion of UvCGBP1 affects hyphal growth, conidiation and various stress response. (a) Colony morphology of the WT, ΔUvCGBP1-33, ΔUvCGBP1-36 and CΔUvCGBP1-33 strains on PSA for 14 days. (b) Colony diameter of the mutant strains on PSA for 14 days. (c) Mycelial biomass of the mutant strains. (d) Hyphal tips stained by CFW. Scale bar = 60 μm. (e) The apical compartment length of the mutant strains. (f) Conidial production of the mutant strains from PSB at 180 rpm for 7 days. (g) Colony growth of the mutant strains on PSA supplied with 0.3 M NaCl, 0.5 M sorbitol, 0.015% H2O2, 0.03% SDS, 0.12 mg/ml CR or 0.12 mg/ml CFW after incubation of 14 days at 28°C. (h) Inhibition rate of colony growth by various stress inducers. Asterisks represent significant differences between the WT and mutants based on LSD at P = 0.05
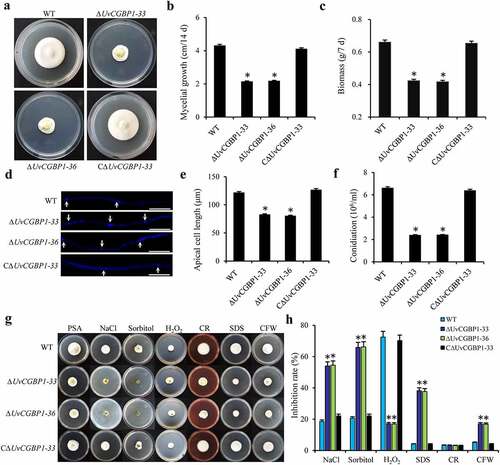
Figure 3. UvCGBP1 is important for invasive growth and virulence. (a) Virulence assays of the WT, ΔUvCGBP1-33, ΔUvCGBP1-36 and CΔUvCGBP1-33 strains on rice spikelets at 21 dpi. (b) Infection observation in inoculated rice spikelets at 1 and 6 dpi by SEM. Scale bars = 20 μm (1 dpi) and 100 μm (6 dpi)
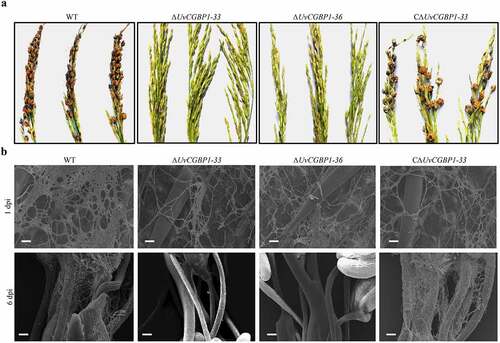
Figure 4. UvCGBP1 regulating virulence might be independent of cutinase genes. (a) Expression of cutinase genes in the WT and ∆UvCGBP1-33 by qRT-PCR relative to β-tubulin. (b) Yeast one-hybrid analysis indicated UvCGBP1 can bind to the promoter of these four cutinase genes (Uv8b_2652, Uv8b_7184, Uv8b_7527 and Uv8b_8131). (c) Colony morphology of the WT and cutinase gene deletion mutants on PSA for 14 days. (d) Virulence assays of the WT and cutinase gene deletion mutants on rice spikelets at 21 dpi. (e) Determination of cutinase activity in 7-day-old hyphae from PSB between the ∆UvCGBP1-33, cutinase gene deletion mutants and WT. Asterisks represent significant differences between WT and mutants based on LSD at P = 0.05
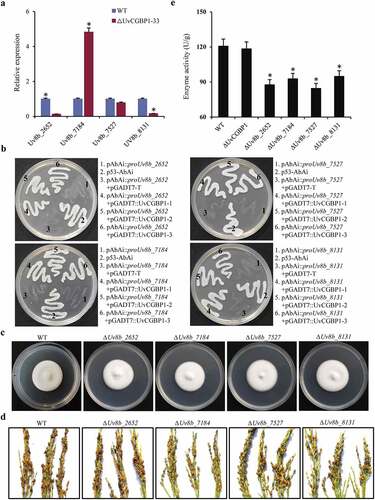
Figure 5. Genome-wide binding sites of UvCGBP1 identified by ChIP-Seq. (a) UvCGBP1 binding peaks and target genes identified by ChIP-Seq. (b) Distribution of UvCGBP1-binding sites. (c) UvCGBP1-binding sites were shown in the genome of U. virens. (d) Functional categories of UvCGBP1-bound genes. (e) KEGG enrichment analysis of UvCGBP1-bound gene-associated ChIP-seq data set. (f) Identification of UvCGBP1 binding motif with MEME (Top six). (g) ChIP-qPCR validated ChIP-seq results using five selected targets. Non-target gene, Uv8b_900 (β-tubulin), was used as a negative control. Asterisk means that UvCGBP1 was significantly enriched on the promoter of tested target gene (P = 0.05)
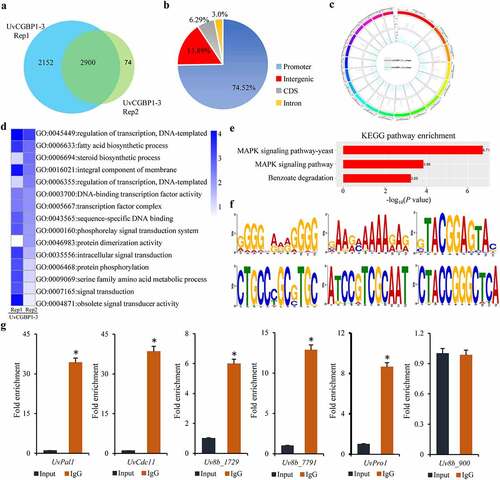
Figure 6. Integrative analysis of RNA-seq and ChIP-seq data. (a) Heatmap and (b) scatter plot indicated up- and down- regulated UvCGBP1-binding target genes. (c) Basic activation/repressive function prediction of UvCGBP1 by BETA software. (d) UvCGBP1-binding target genes significantly enriched in C/AAGGGG motif by MEM-ChIP. (e) EMSA confirmed that UvCGBP1 can bind C/AAGGGG motif. (f,g) Functional categorization of up- and down-regulated UvCGBP1-binding target genes
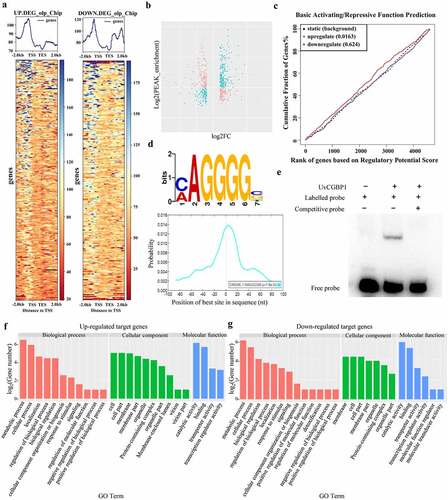
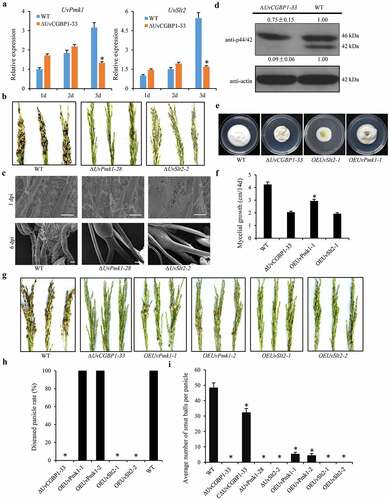
Figure 7. UvCGBP1 regulates virulence through mediating MAPK pathway. (a) Expressions of UvPmk1 and UvSlt2 in the WT and ∆UvCGBP1-33 during the early infection process (1–3 dpi) by qRT-PCR. Asterisks represent significant differences between the WT and mutant based on LSD at P = 0.05. (b) Virulence assays of UvPmk1 and UvSlt2 deletion mutants on rice spikelets at 21 dpi. (c) Infection development of UvPmk1 and UvSlt2 deletion mutants on rice spikelets under SEM at 1 and 6 dpi. Scale bars = 50 μm (1 dpi) and 100 μm (6 dpi). (d) Western blot analysis of proteins extracted from PSB-cultured hyphae of the ∆UvCGBP1-33 and WT using anti-p42/44 antibody. The WT loading set was considered as 1.00, and relative quantified signals of each band were calculated. (e) Colony morphology of UvPmk1 and UvSlt2 over-expression mutants in ∆UvCGBP1-33 and WT on PSA for 14 days. (f) Mycelial growth of ∆UvCGBP1-33, OEUvPmk1-1, OEUvSlt2-1 and the WT on PSA for 14 days. Asterisks represent significant differences between the ∆UvCGBP1-33 and OEUvPmk1-1 based on LSD at P = 0.05. (g) Virulence assay of the over-expression mutants in ∆UvCGBP1-33 on rice spikelets at 21 dpi. (h) Disease incidence in rice panicles. (i) Mean number of smut balls per panicle. Asterisks represent significant differences between the WT and other strains based on LSD at P = 0.05