Figures & data
Figure 1. sRNA23 is a downregulated sRNA in response to iron starvation. (a) Growth of S. suis 05ZYH33 under iron starvation. S. suis were grown in THB + 5% FBS. After 30 minutes of bacterial culture, BIP was added to cultures. The black arrow indicates harvest time point for control and BIP-treated samples. (b) Expression levels of various sRNAs in S. suis 05ZYH33 at the indicated hours of post culture were examined by RT-PCR. (c) RT-PCR detection of three sRNAs of S.suis 05ZYH33 under iron starvation
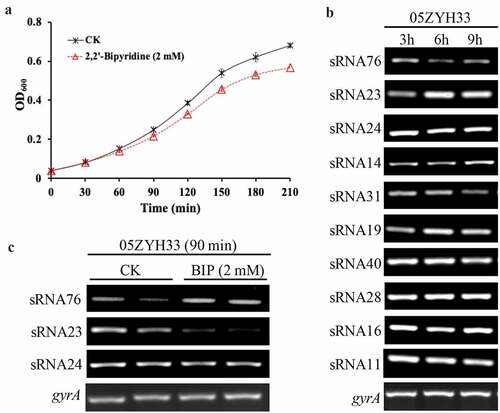
Figure 2. sRNA23 deletion impairs the pathogenicity of 05ZYH33. (a) Detection of mutant strains at the genome level by PCR using Out-F/R primers (Figure S1) respective to genomic DNA bands of WT and sRNA deleted mutants detected by gel electrophoresis. (b) Gram staining results of WT and S. suis mutants under light microscope (magnification, 1,000X). (c) Growth kinetics of 05ZYH33, ΔsRNA23, ΔsRNA24, and ΔsRNA76 cultured in THB + 5% FBS. (d) Survival rate of BALB/c mice infected with WT and mutant S. suis. BALB/c mice (5-week-old, female, ~22 g) were intraperitoneally injected with 0.4 mL (~5 × 108 CFU) S. suis at the late-exponential growth phase cultured in THB + 5% FBS. (e) HE staining showing histopathological changes in mice infected with WT and mutant S. suis for 72 h. Images are representative of three independent experiments
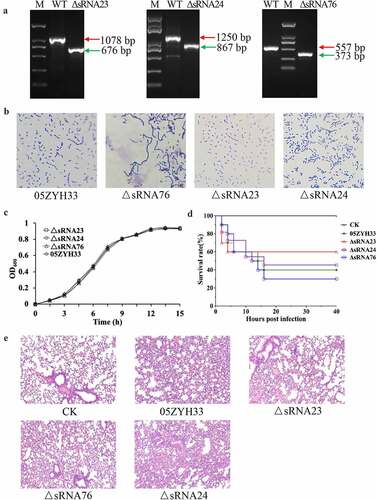
Figure 3. Genomic location and predicted structure of sRNA23. (a) The genomic position of sRNA 23 in S. suis 05ZYH33 was assessed by 5ʹ RACE and 3ʹ RACE. (b) Northern blot analysis for sRNA 23 of 05ZYH33 grown in THB + 5% FBS at exponential phase on formaldehyde denaturing 1.2% agarose gel. Dig-labeled RNA probes were used to detect target sRNA. The RNA marker was cut off and stained by GelStain. (c) Northern blot analysis of sRNA 23 expression at different growth phases (early-log (OD600: ~0.3), mid-log (OD600: ~0.6), and late-log (OD600: ~0.8)) of 05ZYH33. 23s rRNA and 16s rRNA were detected as loading controls by using methyl blue staining. (d) Predicted secondary structure of sRNA 23 by RNAfold
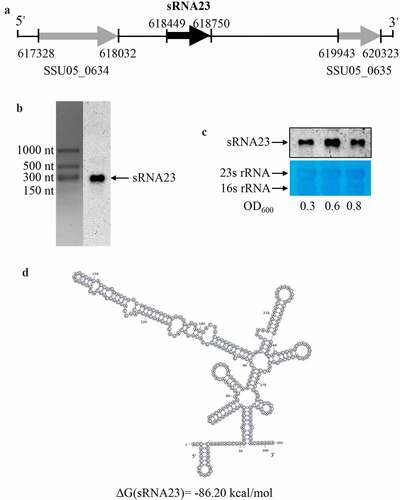
Figure 4. sRNA23 is differentially expressed in a dose-dependent manner under different stress conditions. Northern blot analysis of sRNA23 in S. suis 05ZYH33 strain after treating with different concentrations of (a) bipyridine (0, 1, 2, 4 mM), (b) paraquat (0, 2.5, 5.0, 10 mM), (c) NaCl (0,1, 2, 4 mM) and (d) lysozyme (0, 0.25, 0.5, 1 mg/mL)
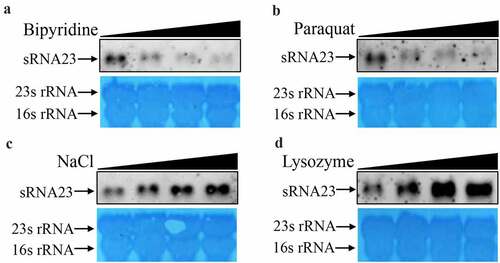
Figure 5. Deletion of sRNA23 has profound effects on cell morphology and biofilm formation. (a) Northern blot analysis for detection of sRNA 23 in 05ZYH33, sRNA 23 deletion strain (ΔsRNA23), and complementation strain (C-∆sRNA23). (b) The OD600 of 05ZYH33, ΔsRNA23, and C-∆sRNA23 cultured in THB + 5% FBS were detected at different time (n = 3). (c) Cell morphology was examined by scanning electron microscope (scale bar, 10 μm). (d) Transmission electron microscope was performed to compare the CPS thickness of 05ZYH33, ΔsRNA23, and C-∆sRNA23. The scale bar indicates 500 nm. (e) The capsular thickness of each strain was quantified using Image J 1.50 software. Data are shown as means ± SD (**P< 0.01). (f) Reduction of biofilm formation by sRNA23 inactivation in 05ZYH33. The spectrophotometer UV-5500 (Shanghai Metash Instruments) was applied to measure optical density at 570 nm (OD570) here. The data are presented as means ± SD (**P< 0.01)
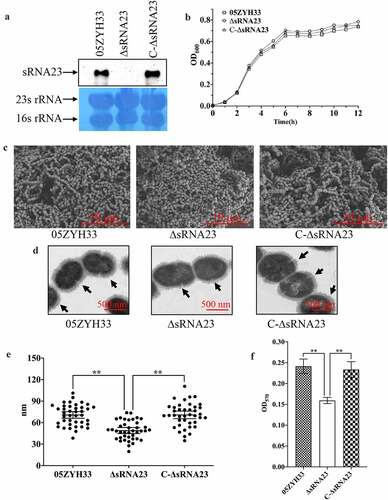
Figure 6. sRNA23 deletion impairs adhesion & hemolytic activity, and enhances susceptibility toward phagocytosis. (a) Relative adherence of the WT strain 05ZYH33, ∆sRNA23 mutants and complementation strain C-∆sRNA23 on HEp-2 cells was analyzed by bacterial adhesion assay (**P< 0.01). (b) Hemolytic activity analysis of S. suis WT strain (05ZYH33), ∆sRNA23, and C-∆sRNA23(**P< 0.01). (c) Survival rate of indicated S suis strains in pig blood (**P< 0.01). (d) Phagocytosis rate of indicated S suis in RAW264.7(**P< 0.01)
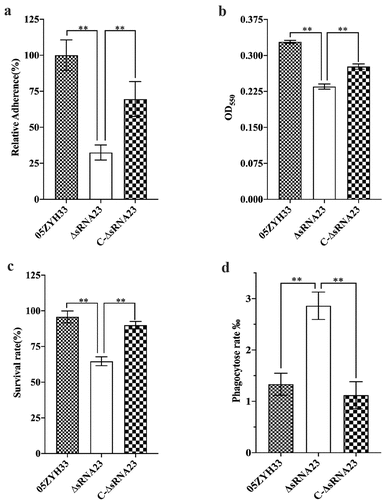
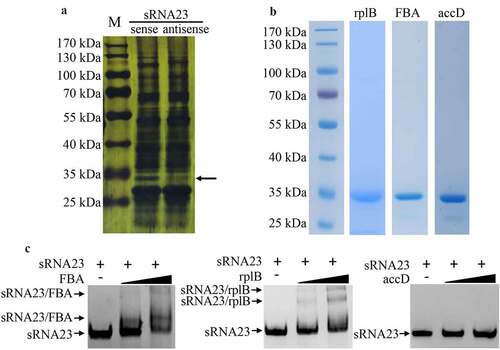
Figure 7. sRNA23 interacts with FBA and rplB. (a) Silver staining of proteins pulled down by sRNA23 sense probes and sRNA23 antisense probes from S. suis 05ZYH33 total protein. The special sRNA23 associated band (arrow) was excised for mass spectrometry. (b) Indicated candidate proteins were expressed in E. coli 21(DE3) and purified by Ni-NTA resin. (c) The binding ability of these purified proteins to sRNA23 was determined by EMSA (FBA 0, 1 μg, 2 μg; rplB 0, 0.5 μg, 1 μg; accD 0, 1 μg, and 2 μg)
Supplemental Material
Download Zip (880.5 KB)Data availability statement
The datasets produced in this study are not available publicly because they are currently private (SRA accession number: PRJNA717999) and are scheduled to be released on 30 December 2021. Any requests to access the datasets should be directed to YC.
