Figures & data
Figure 1. Geographical location of Yunnan and CRFs identified in Southeast Asian countries. The red dot indicates the sampling site
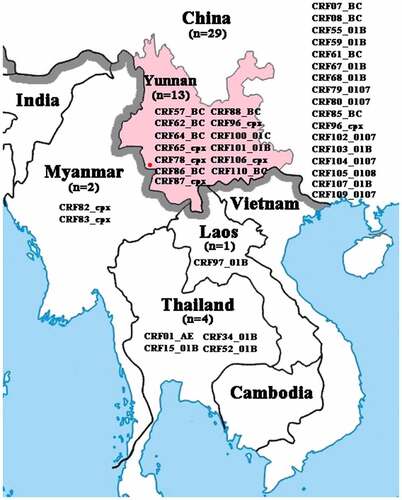
Table 1. Demographic characteristics of 15 HIV-1 infected individuals
Figure 2. The maximum likelihood tree of 15 HIV-1 near full-length genomic sequences from Yunnan, China. All 15 sequences obtained in this study are highlighted with red circles. The blue and red shadows indicate the clades of CRF111_01 C and CRF116_0108, respectively. The red star indicates that 14YN252 belongs to the CRF106_cpx variant
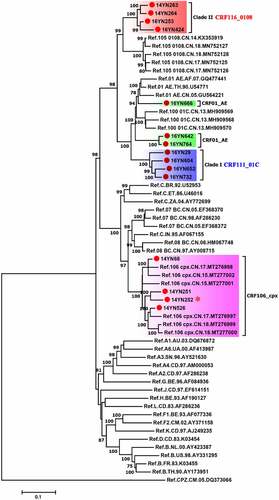
Figure 3. Genomic structural maps for quasispecies of HIV-1 (HXB2:6024–7381) from 14YN251 and 14YN252
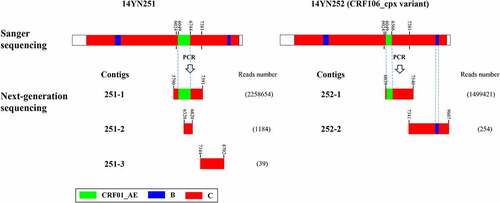
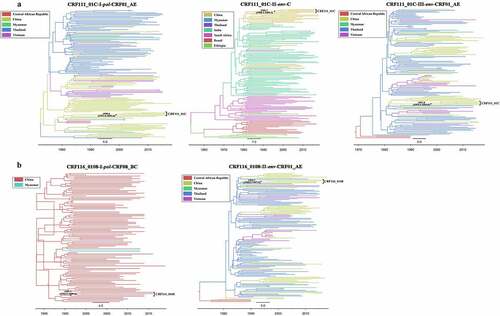
Figure 4. Maximum clade credibility (MCC) trees of the HIV-1 CRF111_01 C and CRF116_0108. (a) The reconstructed MCC tree based on three genomic segments of CRF111_01 C, including the pol region of subtype CRF01_AE origin (3600–4600 nt in HXB2), env region of subtype C origin (7002–7584 nt in HXB2) and env region of subtype CRF01_AE origin (7790–8790 nt in HXB2). (b) The reconstructed MCC tree based on two genomic segments of CRF116_0108, including the pol region of subtype CRF08_BC origin (3200–4200 nt in HXB2) and the env region of subtype CRF01_AE origin (7002–8002 nt in HXB2). Different tree branch colors represent different countries. The black solid dots on the trees indicate the tMRCA of CRF111_01 C and CRF116_0108
Supplemental Material
Download MS Word (2.4 MB)Data availability statement
The datasets presented in this study can be found in online repositories. These data can be found here: https://www.ncbi.nlm.nih.gov/nuccore/MT624743.
