Figures & data
Table 1. Oligo primers used for PCR amplification (restriction enzyme cleavage sites are shown in bold)
Figure 1. Identification of plasma proteins binding to rSpy0136/CEF. A. Recombinant CEF was expressed in E. coli and purified by immobilized metal affinity chromatography (IMAC), and size exclusion chromatography. The purity of rCEF was confirmed by SDS-PAGE. B. Purified rCEF was immobilized on sepharose beads and used for a pull-down experiment with human plasma. (a) Washed beads were loaded on a SDS-PAGE gel. (b) Gel slices extracted for mass spectrophotometry analysis. Mass-spectrometry results identified proteins: (1) rCEF, (2) fibrinogen β-chain, (3) C1r complement protein, C1s complement protein, Fibulin-1, (4) C4BPA, C3 complement protein. (c) Negative control of uncoupled Sepharose beads.
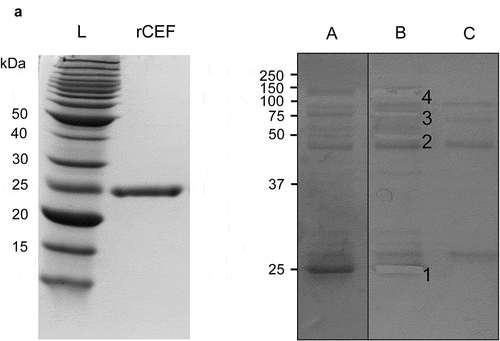
Figure 2. CEF binds to complement proteins. Binding of complement proteins C1r, C1s, C2, C3, C5 and C4BPA was analyzed by ELISA using immobilized rCEF. Proteins were treated or untreated with PNGase F to remove N-linked glycans. Dose-dependent binding was detected using specific primary antibodies and a secondary HRP-conjugated anti-rabbit IgG antibody. Red line: before PNGase F treatment. Black line: after PNGase F treatment. Data represent the mean ± SD of three independent experiments in duplicates.
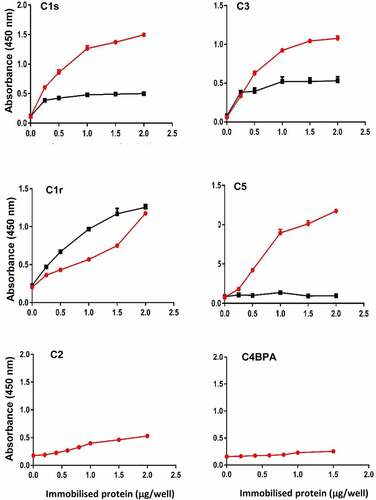
Figure 3. rCEF interferes with complement function. A. Complement-mediated lysis of red blood cells (RBCs). rCEF significantly reduced hemolysis of RBCs in a dose-dependent manner. Staphylococcus aureus SSL7, a known complement inhibitor [Citation45] was used as a positive control. The nonfunctional mutant SSL7-C5−[Citation46] was used as a negative control. Data represent hemolysis as mean ± SD of triplicate A412nm readings. **p < 0.0001, unpaired two-tailed t-test. B. Wieslab human complement activity assay. Activation of individual complement pathways was assessed using the Wieslab® complement system screen kit. rCEF, SSL7 (positive control), and SSL7-C5− (negative control) were tested at a concentration of 2 µM. The interference with the activity of each of the three complement pathways is shown in comparison to the control serum without the added proteins. The C5b-9 complex (membrane attack complex, MAC) was detected with a specific alkaline phosphatase labeled antibody to a neoantigen of the MAC complex. Analyzing the result by one-way ANOVA revealed significant differences (**p < 0.0001) between the marked groups. No significant difference was observed between the negative control and serum-only group. Data are presented as means ± SD for two independent experiments performed in duplicate.
![Figure 3. rCEF interferes with complement function. A. Complement-mediated lysis of red blood cells (RBCs). rCEF significantly reduced hemolysis of RBCs in a dose-dependent manner. Staphylococcus aureus SSL7, a known complement inhibitor [Citation45] was used as a positive control. The nonfunctional mutant SSL7-C5−[Citation46] was used as a negative control. Data represent hemolysis as mean ± SD of triplicate A412nm readings. **p < 0.0001, unpaired two-tailed t-test. B. Wieslab human complement activity assay. Activation of individual complement pathways was assessed using the Wieslab® complement system screen kit. rCEF, SSL7 (positive control), and SSL7-C5− (negative control) were tested at a concentration of 2 µM. The interference with the activity of each of the three complement pathways is shown in comparison to the control serum without the added proteins. The C5b-9 complex (membrane attack complex, MAC) was detected with a specific alkaline phosphatase labeled antibody to a neoantigen of the MAC complex. Analyzing the result by one-way ANOVA revealed significant differences (**p < 0.0001) between the marked groups. No significant difference was observed between the negative control and serum-only group. Data are presented as means ± SD for two independent experiments performed in duplicate.](/cms/asset/d8ed97ec-b1a3-4629-889d-48d5b17b0c0d/kvir_a_2027629_f0003_oc.jpg)
Figure 4. CEF inhibits deposition of MAC and C3b on the surface of S. pyogenes. The deposition of MAC (C5b-9) (a, b) or C3b (c, d) on the surface of S. pyogenes was analyzed by flow cytometry. The bacteria were opsonised with 10% human serum in the absence (Ops S. pyogenes) or presence of 0.5 μM rCEF (Ops S. pyogenes + rCEF). Non-opsonised (Non-ops S. pyogenes) were used as a control. (a, c) The flow cytometry histogram shows an overlay of bacteria incubated with human serum (red), buffer (Orange), or serum + rCEF (blue). A gate has been applied to identify the percentage of C5b-9 or C3b deposition (FITC-A+). Bar graph displaying the percentage of bacterial cells positive for C5b-9 (b) or C3b (d). Analyzing the result by one-way ANOVA revealed significant differences between the groups (**p < 0.0001).
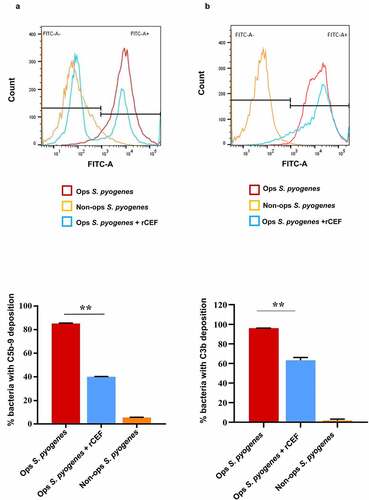
Figure 5. CEF promotes bacterial survival in human blood. (a) Survival rate of S. pyogenes wild-type (WT), S. pyogenes Δcef deletion mutant, and S. pyogenes Δcef:cef complementation strain after incubation with whole human blood. Deletion of cef caused a considerably reduced survival rate compared to the CEF expressing strains. (b) Survival of L. lactis wild-type (WT) and a L. lactis cef gain-of-function mutant in whole human blood. The mutant strain showed an improved survival rate compared to wild-type L. lactis. Survival rates were calculated by dividing the CFU at a given time point by the CFU of the initial inoculum. The data shown are representative of three independent experiments, displayed as the mean ± SD. Significant differences between strains were calculated by Student’s t-test, *p < 0.05, **p < 0.0001.
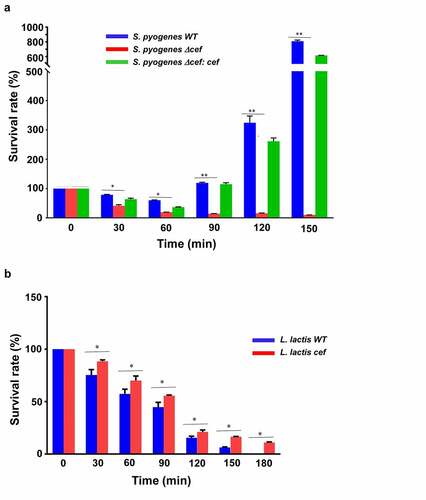
Figure 6. CEF is a crucial virulence factor in a Galleria mellonella wax moth larvae infection model. Antibiotic-free G. mellonella larvae (n = 10) were injected with 20 µl (1.5x106 CFU) of S. pyogenes WT, S. pyogenes Δcef or S. pyogenes Δcef:cef into the lower-left proleg and monitored daily for 5 days. A. Survival of larvae is shown as Kaplan-Meier survival curves. **p < 0.0001 (log-rank test) B. Health index scores for infected larvae. Data shows the mean ± SEM, **p < 0.0001 (2-way ANOVA).
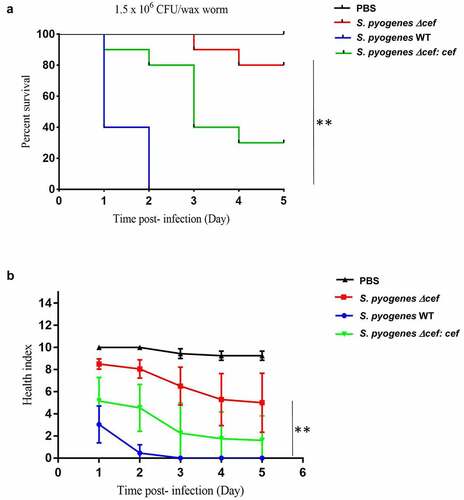
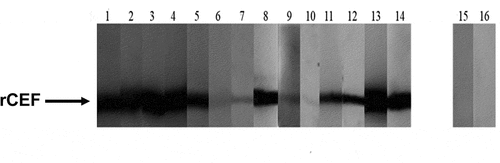
Figure 7. Specific anti-CEF antibodies in serum from patients with S. pyogenes disease. Recombinant CEF (2 μg/lane) was run on a 12% SDS-PAGE gel and blotted onto a nitrocellulose membrane. Individual strips of the Western blot were probed with serum from S. pyogenes invasive disease patients (lanes 1–14) or healthy donors (lanes 15–16) and developed with a secondary HRP-conjugated anti-human Fc antibody.
Supplemental Material
Download Zip (749.2 KB)Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article. Additional data are available from the corresponding author [TP] upon reasonable request.
