Figures & data
Figure 1. Flow cytometry analysis of DHIV3-mCherry infected THP-1 cells and UMAP projection of scRNA seq data from replicate experiment. Panel A) mock infection of PMA activated THP-1 cells. B) PMA activated THP-1 cells infected with DHIV3-mCherry. Absicca, mCherry (Texas Red) emission. Ordinate, GFP (FITC) emission. mCherry positive cells equal approximately 8.5% of the total viable cell population. C) UMAP projection of a duplicate culture (HIVreplicate1) is shown in panel C. Greater than 14,000 different cellular genes were detected in this analysis, including the 9 viral genes and mCherry message originating from DHIV3-mCherry. A semi-supervised two cluster model was adopted, the smaller “Provirus cluster” (cluster A) was 8.1% of the total cell population, approximately equivalent to the percentage of mCherry positive cells from panel B. The two semi-supervised clusters are circled in red. PIC/Bystander cluster is indicated as cluster B. The HIV activity scale presents the Seurat module score that is described in methods. Input data in this analysis included 33,819 PCA entries. Bar codes of the same cells tracked to the Provirus clusters, regardless of whether the clusters were generated using UMAP or Seurat-tSne tools (Fig. S-3).
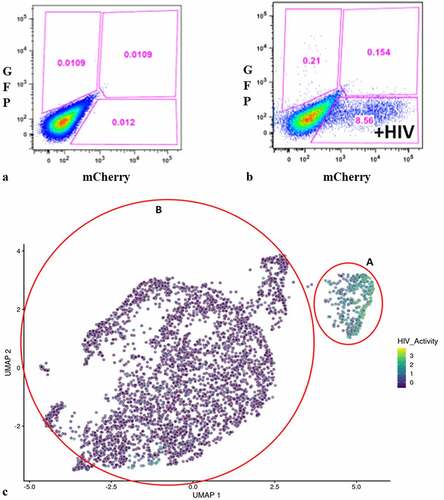
Figure 2. Unsupervised clustering of UMAP shown in . Panel A) shows unsupervised clustering obtained at K equals 10. B) Violin plot of HIV-1 transctipts/cell in the 10 clusters identified at K10 (Scran’s buildSSNGraph using the PCA as input). PIC cells with detectable HIV-1 transcripts, were distributed throughout clusters 1–5, 7 and 9–10. Clusters 6 and 8 contained 372 of the 381 cells included in the semi-supervised Provirus cluster (circled in red). Stipulation of lower K values means that during analysis any one given cell is clustered with a smaller number of cells with similar transcriptomes.
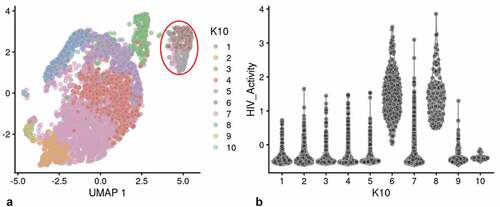
Figure 3. Integrase-inhibitor treatment selectively reduces mCherry positive cells. Panel A) flow cytometry analysis of DHIV3-mCherry infected THP-1 cells, versus viability stain. Abscissa shows viability stain intensity, ordinate shows mCherry intensity. Infected (Provirus), mCherry-producing cells account for approximately 12% of the cell population. Panel B) Same as A except with the addition of 25 nM MK-2048 integrase inhibitor at time of infection. Integrase inhibitor effectively reduces number of mCherry producing cells, without decreasing cell viability.
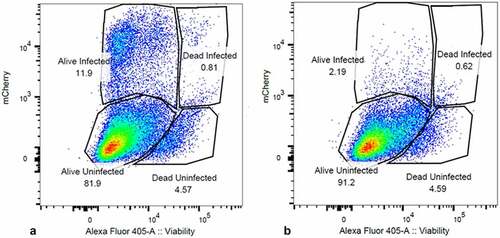
Figure 4. Effect of integrase inhibitor on mCherry, p24, Gag and Vpu protein production in cultures containing DHIV3-mCherry infected cells. MW, molecular weight markers. A) Cells infected with DHIV3-mCherry were purified by FACS sorting based on their expression of mCherry fluorescence. Lane 1, Protein from Control cells; Lane 2, Protein from PIC/Bystander cells; Lane 3, Protein from Provirus cells. Antibody used was goat anti-mCherry, developed with HRP linked anti-goat secondary. mCherry protein was only detectable in sorted Provirus cells. B-E) Lane 1, Control cell protein; Lane 2, protein from DHIV3 infected culture; Lane 3, protein from DHIV3 infected cultures treated with integrase-inhibitor (25 nM MK-2048) as shown above in . B) Lane 2, mCherry protein was readily detectable in protein from cultures containing Provirus cells, with anti-mCherry antibody used in A. Lane 3, a small amount of mCherry signal was detected in MK-2048 treated cultures. C) Lane 2, p24 and Gag precursor proteins visualized with p24 antibody used above in Fig. S-6, and HRP linked anti-mouse secondary antibody. The p24 band in lane 3 is residual from infection as reported in the literature [Citation10]. The presence of precursor proteins in lane 2 shows p24 synthesis in cultures containing Provirus cells. D) Lanes 1 and 2, Control cell protein at 24 and 48 hrs respectively; lanes 3 and 4, protein from DHIV3 infected culture at 24 and 48 hrs respectively; lanes 5 and 6, protein from DHIV3 infected cultures treated with integrase inhibitor (as above) at 24 and 48 hrs respectively. At 24 hrs post infection, we only found both p24 and precursor Gag proteins in the protein samples from DHIV3 infected cells in the absence of integrase inhibitor. At 48 hrs post-infection, in the absence of integrase inhibitor, the amounts of detectable p24 and Gag proteins were dramatically increased from levels at 24 hrs post infection. As seen initially (Panel C), some p24 protein was detectable in integrase inhibitor-treated cultures at 24 hrs post infection, however, Gag is not detectable at this time. At 48 hrs post infection in the integrase inhibitor treated cultures, some Gag protein does become detectable, reflecting production in cells that escaped complete integrase inhibition. This is in agreement with our flow cytometry analysis that showed suppressed, but still detectable numbers of mCherry positive cells in the integrase inhibitor treated cultures (). The Gag precursor proteins only appear in the integrase inhibitor treated culture proteins 48 hrs after treatment. All antibodies, sources and dilutions are provided in Methods. E) Lane 2, Vpu detected with rabbit antibody, visualized using HRP linked anti-rabbit secondary. The resolution of the image is slightly compromised due to the small size of Vpu protein.
![Figure 4. Effect of integrase inhibitor on mCherry, p24, Gag and Vpu protein production in cultures containing DHIV3-mCherry infected cells. MW, molecular weight markers. A) Cells infected with DHIV3-mCherry were purified by FACS sorting based on their expression of mCherry fluorescence. Lane 1, Protein from Control cells; Lane 2, Protein from PIC/Bystander cells; Lane 3, Protein from Provirus cells. Antibody used was goat anti-mCherry, developed with HRP linked anti-goat secondary. mCherry protein was only detectable in sorted Provirus cells. B-E) Lane 1, Control cell protein; Lane 2, protein from DHIV3 infected culture; Lane 3, protein from DHIV3 infected cultures treated with integrase-inhibitor (25 nM MK-2048) as shown above in Figure 3. B) Lane 2, mCherry protein was readily detectable in protein from cultures containing Provirus cells, with anti-mCherry antibody used in A. Lane 3, a small amount of mCherry signal was detected in MK-2048 treated cultures. C) Lane 2, p24 and Gag precursor proteins visualized with p24 antibody used above in Fig. S-6, and HRP linked anti-mouse secondary antibody. The p24 band in lane 3 is residual from infection as reported in the literature [Citation10]. The presence of precursor proteins in lane 2 shows p24 synthesis in cultures containing Provirus cells. D) Lanes 1 and 2, Control cell protein at 24 and 48 hrs respectively; lanes 3 and 4, protein from DHIV3 infected culture at 24 and 48 hrs respectively; lanes 5 and 6, protein from DHIV3 infected cultures treated with integrase inhibitor (as above) at 24 and 48 hrs respectively. At 24 hrs post infection, we only found both p24 and precursor Gag proteins in the protein samples from DHIV3 infected cells in the absence of integrase inhibitor. At 48 hrs post-infection, in the absence of integrase inhibitor, the amounts of detectable p24 and Gag proteins were dramatically increased from levels at 24 hrs post infection. As seen initially (Panel C), some p24 protein was detectable in integrase inhibitor-treated cultures at 24 hrs post infection, however, Gag is not detectable at this time. At 48 hrs post infection in the integrase inhibitor treated cultures, some Gag protein does become detectable, reflecting production in cells that escaped complete integrase inhibition. This is in agreement with our flow cytometry analysis that showed suppressed, but still detectable numbers of mCherry positive cells in the integrase inhibitor treated cultures (Figure 3). The Gag precursor proteins only appear in the integrase inhibitor treated culture proteins 48 hrs after treatment. All antibodies, sources and dilutions are provided in Methods. E) Lane 2, Vpu detected with rabbit antibody, visualized using HRP linked anti-rabbit secondary. The resolution of the image is slightly compromised due to the small size of Vpu protein.](/cms/asset/d5c9b3f7-2837-447b-b150-912fb7ad0bc3/kvir_a_2031583_f0004_b.gif)
Figure 5. UMAP analysis of integrase-inhibitor treated DHIV3-mCherry infected THP-1 cells. Experiment performed as shown in , with 25 nM MK-2048 added at the time of DHIV3 addition. Data were analyzed identically to data shown in . Panel A) Feature plot showing the distribution of cells containing HIV-1 transcript, generated as described. Panel B) K10 unsupervised clustering generated 7 clusters (Scran’s buildSSNGraph using the PCA as input). HIV-1 transcripts were distributed equally throughout all of them. No cluster corresponding to the “Provirus” cluster detected in was detected, regardless of K value used (see Fig. S-8). These data agree with the concept that integrase inhibitors selectively target and reduce the number of Provirus cluster cells.
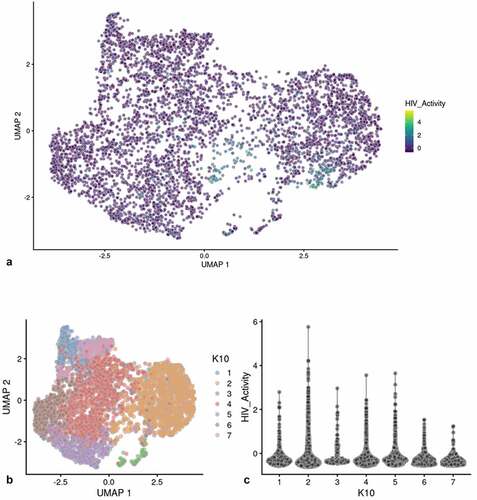
Table 1. Hallmark analysis of gene pathways up or down regulated detected in Provirus versus PIC/Bystander cluster cells (respectively). GSEA Hallmark analysis (fgsea R package) of metabolic pathways negatively or positively regulated (p < 0.1) in the Provirus cluster transcriptome versus the PIC/Bystander cluster transcriptome. Pathways up-regulated in Provirus cells include E2F, Myc targets, G2-M checkpoint, spermatogenesis and oxidative phosphorylation. Down-regulated pathways identified are more numerous, but notably included TNFα signaling via NFkB, inflammatory response genes, apoptosis and interferon γ response. The pairwise T-Tests function from Scran was used to determine the significant of genes between groups. The significant DGE subsets were used for all comparisons
Figure 6. Unsupervised clustering of HIVrepeat2. Panel A) shows unsupervised clustering obtained at K equals 10. Panel B) Violin plot of HIV-1 transctipts/cell in the 10 clusters identified at K10 (Scran’s buildSSNGraph using the PCA as input). PIC cells with detectable HIV-1 transcripts were distributed throughout clusters 1, 2 and 4–10. Cluster 3 contained 135 of the 227 cells in the semi-supervised Provirus cluster (circled in red).
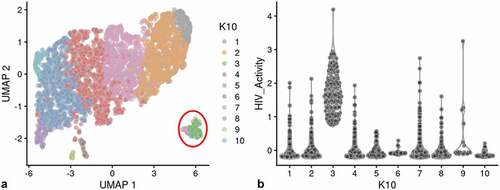
Figure 7. Dotplot of genes associated with M0, M1 and M2 differentiation states. The expression of M0, M1 or M2 marker genes in Provirus cells was not found to differ from those found in Control cells. The overall gene expression pattern did not change appreciably with HIV integration indicating that there was not a change in the differentiated THP-1 state with the viral infection. Minor changes are observed in the PIC and Bystander cells. These conditions were found to have higher overall levels of expression of the MAFB (M0) and IL1B, HLA-DRB1, and CD68 (M1) differentiation marker genes. This analysis shows relative upregulation M0 and M1 markers transcripts in PIC and Bystander cells when compared to Control or Provirus cells.
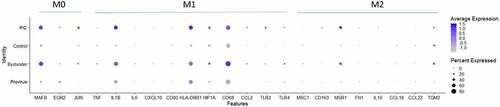
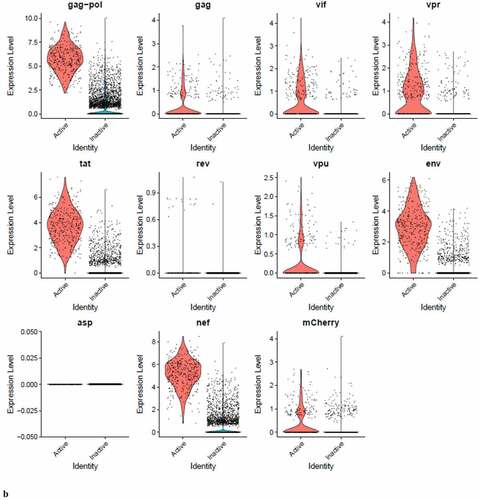
Figure 8. The Distribution of HIV-1 transcripts throughout Provirus and PIC/Bystander clusters. Panel A) Feature plot showing the distribution of cells from UMAP in that contain detectable DHIV3-mCherry transcripts. As described above, these UMAP projections were made with Seurat’s FeaturePlot function. They are colored by the expression of individual genes (UMAP projection colored by walktrap, normalized log2 values). ASP is a negative control, bacterial gene transcript sequence. B) Violin plots of DHIV3-mCherry transcript/cell in cells from the Provirus and PIC clusters showing transcript level and cell number. The provirus cluster contained transcriptomes of 371 cells, the number of PIC cells in the PIC/Bystander cluster was 569 cells, thus the Provirus/PIC cell number ratio was 0.65. The plots were made with Seurat’s VlnPot function. They show normalized log2 transcript levels. Two patterns of transcript distribution are evident. The first pattern is seen with gag-pol, tat, env, and nef, in which relatively high numbers of cells in the PIC/Bystander cluster express the transcripts, with the transcripts being detected in fewer numbers of Provirus cluster cells. The second pattern is seen with gag, vif, vpr, rev, vpu, and mCherry, in which relatively equal absolute numbers of cells in the Provirus and PIC/Bystander clusters are detected with the transcript sequences, remembering that there are more PIC cells than Provirus cells. The relative transcript loads per PIC cell versus the Provirus cells overlap. Negative control sequence (asp) shows no distribution.
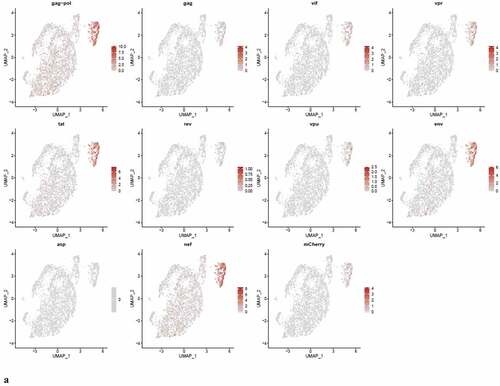
Figure 9. The Distribution of HIV-1 transcripts throughout Provirus and PIC/Bystander clusters in HIVrepeat2. Violin plots of DHIV3-mCherry transcript/cell in cells from the Provirus and PIC clusters showing transcript level and cell number. As described above, these were made with Seurat’s VlnPot function. They show normalized log2 transcript levels. The two patterns of transcript distribution observed in HIVrepeat1 are evident. The first pattern is seen with gag-pol, tat, env and nef, in which higher numbers of cells in the PIC/Bystander cluster detectably express the transcripts. The second pattern is seen with gag, vif, vpr, vpu, and mCherry, in which fewer Provirus or PIC cluster cells are detected expressing the transcripts, but those cells expressing the transcripts are doing so at slightly higher average levels of transcripts per cell. It is difficult to compare transcript loads in the Provirus cluster cells to the results in HIVrepeat1 () due to the lower number of Provirus cells detected in this HIVrepeat2 experiment. In this experiment, the ratio of Provirus cells to PIC cells was 0.17. Nevertheless, the relative patterns observed in HIVrepeat1 are observed here. Following Seurat QC, no Provirus cells expressing rev were detected. Negative control sequence (asp) shows no distribution.
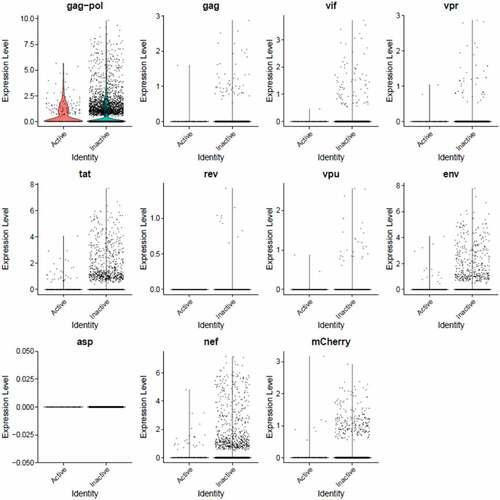
Figure 10. Psupertime analysis of Control, PIC/Bystander, and Provirus cell transcriptomes. Psupertime analysis is a supervised pseudotime approach that explicitly uses sequential labels as input. It uses a regression-based model that acknowledges the cell labels to identify genes relevant to the process. Panel A) one thousand randomly Control (WT), PIC/Bystander (PIC/B), and Provirus (Pro) cell transcriptomes were randomly selected and analyzed. Imposition of identity revealed a pseudo-evolution of Control to PIC/Bystander to Provirus cell transcriptomes. Panel B) distribution of HIV-1 transcripts through these clusters agrees with results shown in , showing no bias toward early or later gene transcripts.
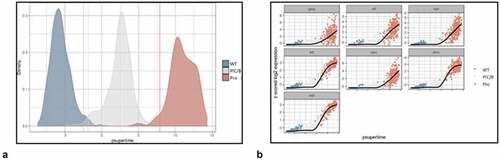
Table 2. Transcription Factor Targeting analysis of DGE contrasting PIC/Bystander and Provirus cells. TFT analysis (GSEA with the fgsea R package and the C3 collection from msig) suggests that at least 3 transcription factor families control the transition from PIC/Bystander transcriptomes to Provirus cluster transcriptomes. These are E2F, NFkB and AP1 family promoter proteins. In particular, increased E2F regulated transcription appears to correspond with the transition to production of viral proteins. The pseudo-transition from Control to PIC/Bystander is characterized by a down regulation of E2F family regulated transcripts and up regulation of NFkB and AP1 regulated transcripts Appendix III. In comparing Provirus to PIC/Bystander transcriptomes, E2F family promoted transcripts are up regulated, while NFkB and AP1 transcription products are down regulated. Comparing Provirus to Control transcriptomes shows that overall Provirus cells have increased E2F regulated transcripts and decreased NFkB transcripts (with no significant change detected in AP1 regulation)
Figure 11. Western blot analysis for phospho- Rb or IkB in protein from mCherry negative versus mCherry positive cells. Cells infected with DHIV3-mCherry were purified by FACS sorting based on their expression of mCherry fluorescence. Lane 1, Protein from Control cells; Lane 2, Protein from PIC/Bystander cells; Lane 3, Protein from Provirus cells. Phospho-Rb (Phospho-T821 Rb antibody) was used to quantify Rb pocket phosphorylation, anti-Rb control antibody was used to quantify Rb protein levels relative to actin (visualized with beta-actin antibody). PIC/Bystander cells show the lowest level of Rb phosphorylation, Provirus show the highest, in close agreement with Transcription Factor Targeting results. Phospho-IkB S32 antibody was used to quantify activated IkB. Control cells show the lowest level of IkB phosphorylation, no difference was detectable between Provirus and PIC Cluster cells.
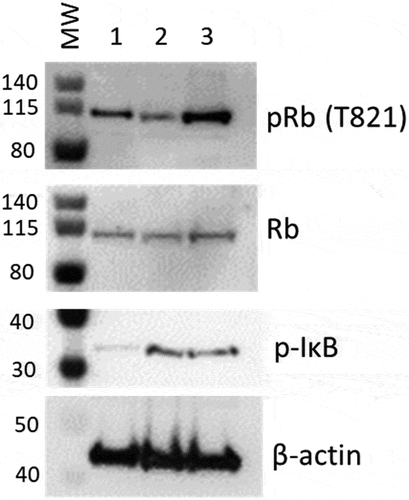
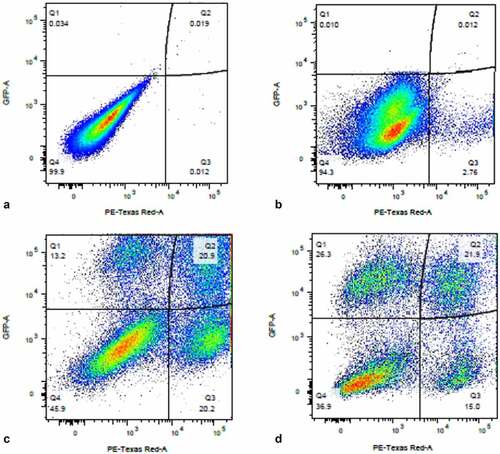
Figure 12. Sequential infection of THP-1 cells with DHIV3-mCherry followed 24 hrs later with GFP DHIV3. Abscissa, mCherry signal, Ordinate, GFP signal. Provirus cluster, mCherry positive, cells were 2 to 5 times more likely to make HIV-1 encoded GFP protein upon the second infection than PIC/Bystander cells upon the second infection. Panel A) time equal 0 hrs; addition of DHIV3-mCherry. Panel B) time equal 24 hrs; addition of DHIV3-GFP. Panel C) time equals 48 hrs after DHIV3-mCherry addition, 24 hrs after DHIV3-GFP addition. Panel D) time equals 72 hrs after DHIV3-mCherry addition, 48 hrs after DHIV3-GFP addition. The percentage of mCherry cells also producing GFP, compared to cells producing mCherry only, is always 2 to 5 times higher than the percentage of cells making only GFP, compared to those cells not producing either mCherry or GFP.
Supplemental Material
Download MS Word (12.1 MB)Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials. The data that support the findings of this study are openly available in “figshare” at the following dois:
Appendix I - https://doi.org/10.6084/m9.figshare.16834633.v1
Appendix II - https://doi.org/10.6084/m9.figshare.16834639.v1
Appendix III - https://doi.org/10.6084/m9.figshare.16834636.v1
Appendix IV - https://doi.org/10.6084/m9.figshare.16834663.v1