Figures & data
Figure 1. Growth characteristics of E. faecalis and its Zn-deficient mutants under Zn-restricted conditions. (a) Schematic of the gene locus of the Zn transport system in E. faecalis OG1RF core genome. Growth curves of E. faecalis wild type (WT) and its isogenic mutants in BHI (b), BHI supplemented with 100 µmZnSo4 (c), 10 µm TPEN (d), combination of TPEN and ZnSo4 (e), TPEN and MnSo4 (f) or TPEN and Fe SO4(g). In (b) and (d-f), data points represent the average of nine biological replicates. Finally, the growth curve of E. faecalis wild type (WT) and genetically complemented ∆adc mutants in BHI supplemented with 10 µm TPEN (h). In (c) and (g), data points represent the average of six biological replicates. Error bar represents the standard error of margin (SEM). Statistical analysis was performed using simple linear regression ofthe exponential growth phase, and slope of each mutant’s growth kinetics was compared with that of the parent strain.
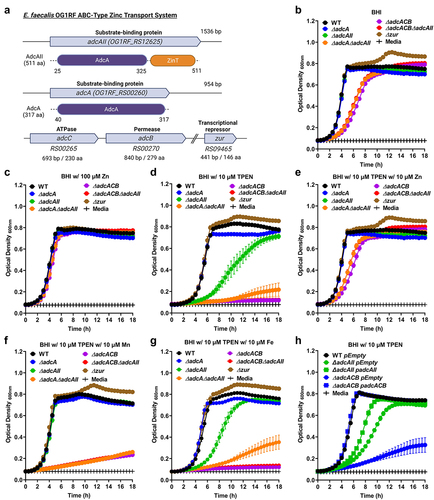
Figure 2. Growth characteristics of E. faecalis and its Zn-deficient mutants in the presence of calprotectin. Growth curves of E. faecalis OG1RF WT and its isogenic mutants in BHI supplemented with CP buffer media (a) and 150 µg ml −1 of WT hCP (b) or hCp∆mn-tail (c). Data points represent the average, and error bar represents the standard error of margin (SEM) of at least six biological replicates. Statistical analysis was performed using simple linear regression of exponential growth phase, and the slope of each mutant’s growth kinetics was compared with that of the parent strain.
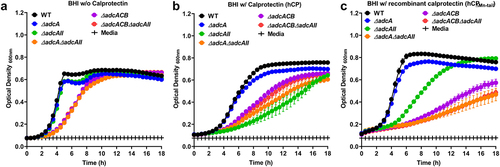
Figure 3. Intracellular Zn quantification and transcriptional profiles of E. faecalis and its Zn-deficient mutants. (a) ICP-OES quantifications of intracellular Zn of mid-log grown E. faecalis OG1RF WT and derivatives grown in BHI and BHI supplemented with 7.5 µm TPEN. Data points represent five biological replicates. Statistical analysis was performed using two-way ANOVA with Dunnett’s multiple comparison test. ** p ≤0.01, *** p ≤0.001, and **** p ≤0.0001. (b) Comparison of reversed transcribed cDNA copy of adcA and adcAII in E. faecalis OG1RF and the ∆zur mutant grown for 1 hour in BHI, BHI with 30 µm TPEN, and BHI with 4 mM ZnSo4. Data points represent the average of six biological replicates. Statistical analysis was performed using the unpaired t-test with Welch’s correction. * p ≤.05. Error bars represent the standard error of margin (SEM).
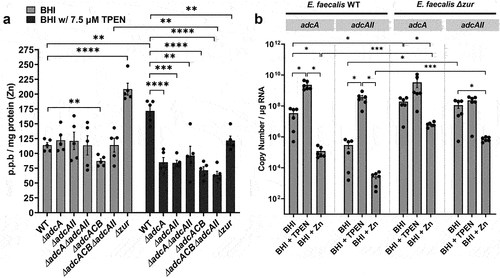
Figure 4. Growth and survival of E. faecalis in serum and urine. Colony-forming units (CFU) of E. faecalis OG1RF WT and its mutants incubated in pooled (a) human serum or (b) serum supplemented with 500 µm ZnSo4 and pooled human urine (d). (c) CFU counts of E. faecalis OG1RF WT, its mutants, and genetic complemented mutants after 24 hours of incubation in pooled human serum. In (a-d), data points represent the average and error bar represents the standard error of margin (SEM) of nine biological replicates. Statistical analysis was performed using one-way ANOVA with Welch’s correction. *** p ≤0.001 and **** p ≤0.0001.
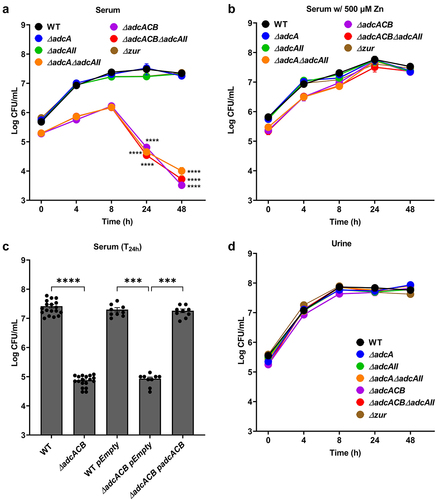
Figure 5. Bright-field microscopic images of E. faecalis and its indicated mutants. Images shown are representative of 10 images that are acquired from one biological sample from each strain grown in BHI, respectively, and imaged at 100x magnification. Black bars represent five microns in length.
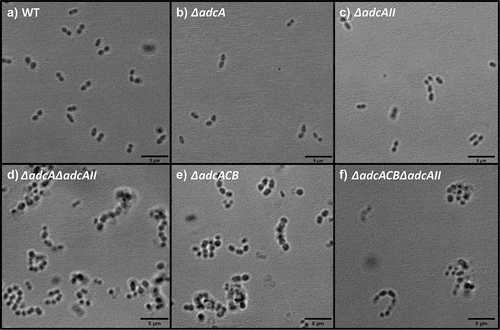
Figure 6. Characterization of E. faecalis virulence traits at the cell surface interface. (a) Biofilm biomass quantification of E. faecalis OG1RF WT and its mutants grown in BHI for 24 hours. Statistical analysis was performed using One-way ANOVA with Welch’s correction. (b-e) Final growth yields of E. faecalis OG1RF WT and its mutants after 24 hours of incubation in BHI supplemented with 2-fold increasing concentrations of (b) ampicillin, (c) daptomycin, (d) bacitracin, and (e) vancomycin. Data points represent the average of nine biological replicates. Statistical analysis was performed using One-way ANOVA with Welch’s correction. * p ≤0.05, *** p ≤0.001, and **** p ≤0.0001. Error bars represent the standard error of margin (SEM).
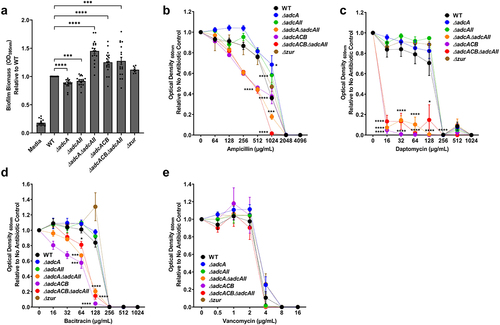
Figure 7. Virulence of E. faecalis in different animal models. (a) Percentage survival of G. mellonella larvae 96 hours post-infection with E. faecalis WT or indicated mutants. Each curve represents a group of 15 larvae injected with ~1 x 105 CFU of selected E. faecalis strain. Data points represent the average of 6 biological replicates. Statistical analysis was performed using the log-rank (Mantel-Cox) test. (b) Total CFU recovered after 48 hours from spleen, peritoneal wash, and catheter of mice infected with 2 × 108 CFU of bacteria. (c) Total CFU recovered after 24 hours from bladder, kidney, spleen, and catheter of mice infected with 1 × 107 CFU of WT or indicated mutants. In (b and c), ten mice were infected with two biological replicates and data points shown were a result of using the ROUT outlier test. The black line represents the median. Statistical analysis was performed using the Mann-Whitney test. * p ≤0.05, ** p ≤0.01, *** p ≤0.001, and **** p ≤0.0001. The dashed line represents the limit of detection (LOD = 50 CFUs).
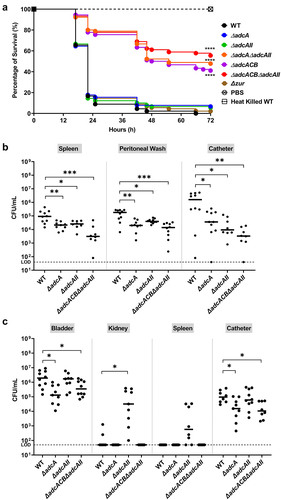
Table 1. Strains and plasmids used in this study
Table 2. Primers used in this study
Supplemental Material
Download Zip (4.4 MB)Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary materials. Vectors and strains created from this study will be available from the corresponding author upon reasonable request.
