Figures & data
Table 1. Fusarium verticillioides strains used in this study
Figure 1. Expression and subcellular localization of FvAtg4 and FvAtg8 in F. verticillioides conidia (a), germ tubes (b), and hyphae (c). F. verticillioides strains expressing the FvAtg4-GFP or GFP-FvAtg8 fusion protein constructs and stained with CMAC were examined by epifluorescence microscopy. AG4-N1 carried the FvAtg4-GFP fusion protein in the ΔFvAtg4 background, and AG8-N2 carried the GFP- FvAtg8 fusion protein in the ΔFvAtg8 background. as a control, GFP was expressed from constructs under the control of the native promoters of FvATG4 and FvATG8. the GFP fluorescence in the resulting transformants, AG4-W2 and AG8-W4, was dispersed throughout the cytoplasm. White arrows indicate the punctate structures. BF=Bright Field. Scale bars = 5 μm.
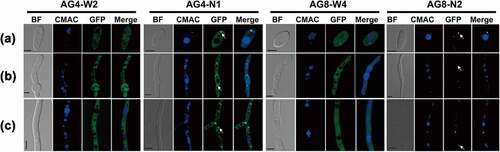
Figure 2. Assay for autophagy defects in the ΔFvAtg4 and ΔFvAtg8 mutants. (a) the vacuoles were filled with autophagosomes in the wild-type 7600 strain, while no autophagic bodies could be observed in the vacuoles of the ΔFvAtg4, ΔFvAtg8 and ΔFvAtg4-8 deletion mutants after growth in MM-N medium supplemented with 1 mM PMSF for 8 h. White arrows indicate the autophagic bodies. “V” is vacuole and “C” is cytoplasm. Scale bar = 5 μm. (b) Organelles and autophagic bodies in the wild-type 7600, ΔFvAtg4, ΔFvAtg8, and ΔFvAtg4-8 mutant strains were observed under starvation conditions. White arrows indicate the autophagic bodies. Scale bar = 0.5 μm. (c) W-AG8 and AG4-R8 carried the GFP- FvAtg8 fusion protein into wild type 7600 or the ΔFvAtg4 background, respectively. Strains were cultured in liquid YEPD and transferred into liquid MM-N medium. Mycelia stained with CMAC were observed using epifluorescence microscopy. BF=Bright Field. Scale bar = 10 μM. (d) GFP-FvAtg8 degradation was verified using an immunoblot assay with anti-GFP antibody in the wild-type 7600 and ΔFvAtg4 strains. Mycelia were harvested at each time point indicated during nitrogen starvation. GAPDH was used as a reference protein.
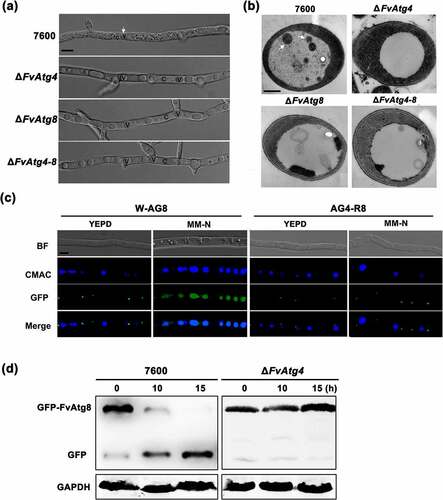
Figure 3. The FvAtg4 protein interacts with the FvAtg8 protein in F. verticillioides. (a) Serial dilutions of yeast cells (cells/ml) transformed with the bait and prey constructs indicated on the left were assayed for growth on SD-Leu-Trp-His plates. a pair of plasmids, pGBKT7–53 and pGADT7, was used as the positive control. a second pair of plasmids, pGBKT7-Lam and pGADT7, was used as the negative control. (b) Colocalization of FvAtg4-red fluorescent protein (FvAtg4-RFP) with green fluorescent protein-FvAtg8 (GFP-FvAtg8) in F. verticillioides. the pair of plasmids FvAtg4-RFP and GFP-FvAtg8 were cotransformed into the wild-type 7600 strain, and the resulting transformant was examined for GFP and RFP signals. (c) Effects of FvATG4 deletion on the cellular movements of GFP-FvAtg8 during conidial germination. Germinated conidia of the wild-type 7600 strain and the FvATG4 deletion mutant expressing the GFP-FvAtg8 fusion construct were stained with CMAC and examined by confocal epifluorescence microscopy. Red arrows indicate the relative position of GFP-FvAtg8 at each time point. BF=Bright Field. Scale bars = 5 μM.
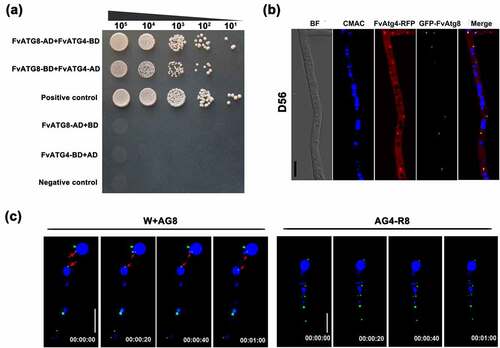
Figure 4. Involvement of FvAtg4 and FvAtg8 in lipid turnover in F. verticillioides. (a) Mycelia of the wild-type 7600 strain, ΔFvAtg4, ΔFvAtg8 and ΔFvAtg4-8 were grown in liquid MM medium and stained with Nile red or BODIPY to observe the lipid droplets in the hyphae. Arrows indicate lipid droplets in the enlarged views. BF=Bright Field. Scale bars = 10 μm. (b) Lipid droplet sizes of the wild-type 7600 strain, ΔFvAtg4, ΔFvAtg8 and ΔFvAtg4-8 were measured. (c) the amount of triglycerides in each strain was determined using a commercially available enzymatic assay kit. Error bars in each column indicate the standard error of three independent experiments. Different letters indicate a significant difference (P < .05).
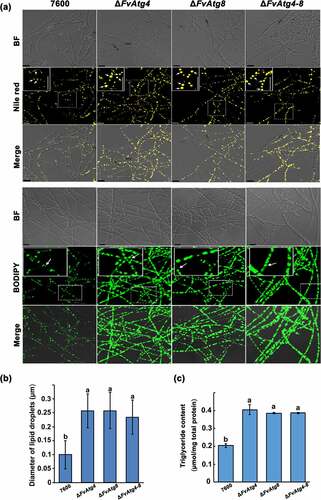
Figure 5. Effects of FvATG4 and FvATG8 deletion on mycelial growth and pigmentation in F. verticillioides. (a) F. verticillioides strains were grown on CM, PDA and MM media at 25°C for 4 d. (b) Effect of four inhibitors on the melanin biosynthesis of the wild-type 7600 strain, ∆fvatg4 and ∆fvatg8. Each strain was incubated in liquid MM and treated with 0.2 mM sodium azide (inhibits laccase in the Raper-Mason pathway), 50 μg/mL tricyclazole (inhibits the dehydrogenation reaction in the DHN pathway), 50 μg/mL mesotrione (inhibits the HGA pathway), or 0.5 mM arginine (inhibits tyrosinase inhibitors in the Raper-Mason pathway). (c) Relative expression of two laccase genes, FVEG_04196 and FVEG_13405, in wild-type F. verticillioides 7600, ∆FvAtg4, ∆FvAtg8 and the ∆FvAtg4-Atg8 double mutant. the expression levels of the laccase genes in ∆FvAtg4, ∆FvAtg8 and ∆FvAtg4-Atg8 are the relative amounts of mRNA in the wild-type progenitor strain. (d) Relative expression of the FvBIK genes in the wild-type 7600 strain and the ∆fvatg4, ∆fvatg8 and ∆fvatg4–8 deletion mutants. Error bars in each column indicate the standard error from three separate experiments. Different letters indicate a significant difference (P < .05).
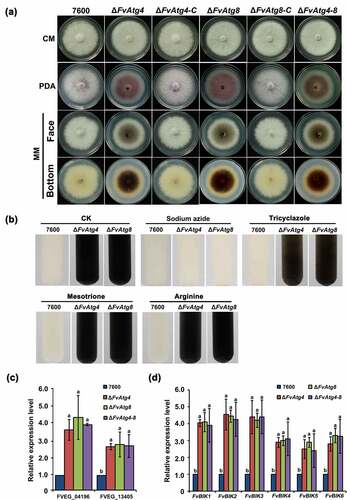
Figure 6. FvAtg4 and FvAtg8 are required for full virulence in F. verticillioides. (a) Disease symptoms of the wild-type 7600 strain, ΔFvAtg4, ΔFvAtg8, ΔFvAtg4-8, ΔFvAtg4-C and ΔFvAtg8-C on maize stalks 15 d after inoculation and (b) on wounded maize kernels 7 d after inoculation. (c) Lesion areas of longitudinally dissected maize stalks infected with F. verticillioides measured after 15 d. (d) Ergosterol production in each strain on maize kernels. Endoglucanase activity (e) and xylanolytic enzyme activity (f) in the wild-type 7600 strain, ΔFvAtg4, ΔFvAtg8, and ΔFvAtg4-8. Error bars denote the standard error of three separate experiments. Different letters indicate a significant difference (P < .05).
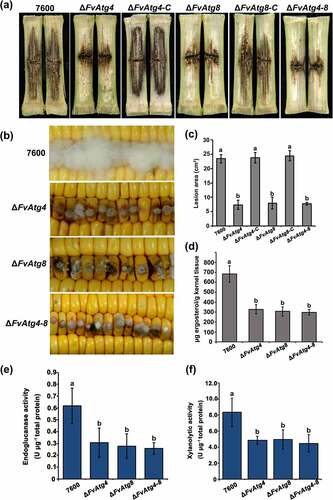
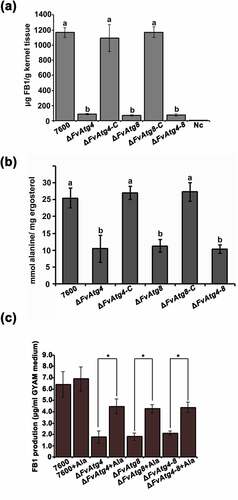
Figure 7. Effects of the FvATG4 and FvATG8 deletions on FB1 biosynthesis in F. verticillioides. (a) The amounts of FB1 (per g maize kernel tissue) produced in infected maize kernels by the F. verticillioides strains 21 d after inoculation. (b) the amount of alanine (per mg fungal ergosterol) produced by each strain after growth in GYAM medium at 25°C for 7 d. (c) Effect of alanine on the FB1 production of the wild-type and the mutants incubated in GYAM medium. Error bars in each column indicate the standard error of three independent experiments. Different letters indicate a significant difference (P < .05).
Supplemental Material
Download MS Word (6.7 MB)Data availability statement
All relevant data are within the manuscript and its Supporting Information files.
