Figures & data
Figure 1. EHEC hemolysis regulation by CsrA.
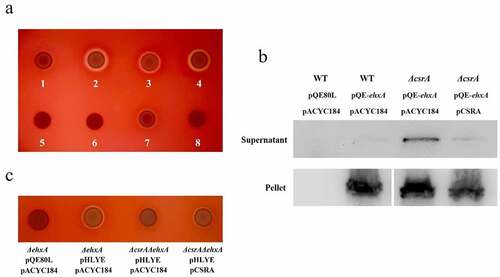
Figure 2. Overview of ehxCABD (EHEC), hlyCABD (UPEC), and hlyE (EHEC) operons.

Figure 3. CsrA-Dependent regulation of ehxB-lacZ, hlyE-lacZ, and ehxA-lacZ translational fusion in vivo.
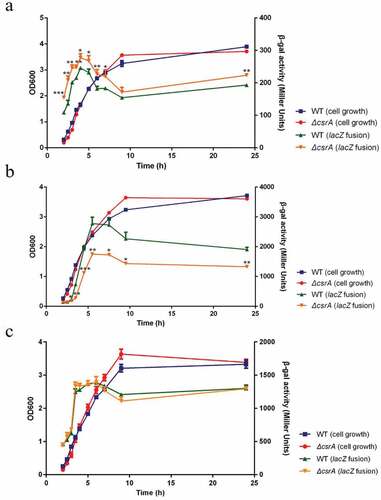
Figure 4. RNA mobility shift assays using various concentrations of purified 6hisCsrA on EHEC.
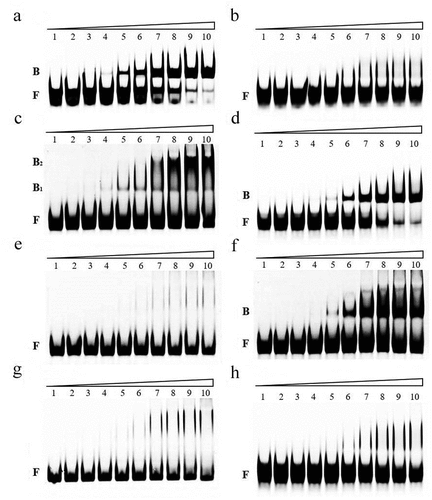
Figure 5. Effect of site-directed mutation in potential binding sites in the expression of lacZ translational fusion in vivo.
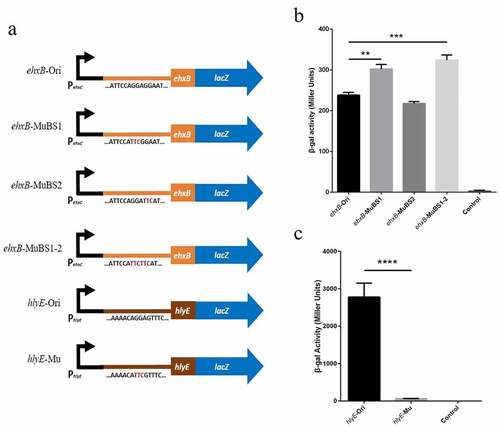
Figure 6. Stability of ehxB and hlyE mRNA as affected by CsrA in EHEC.
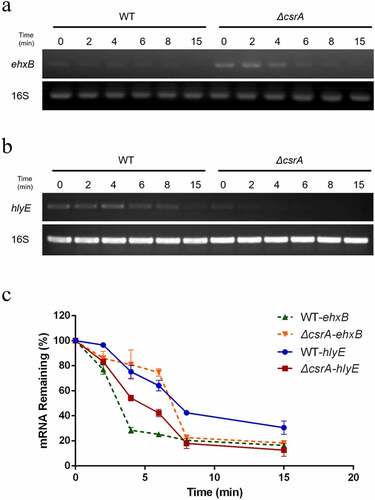
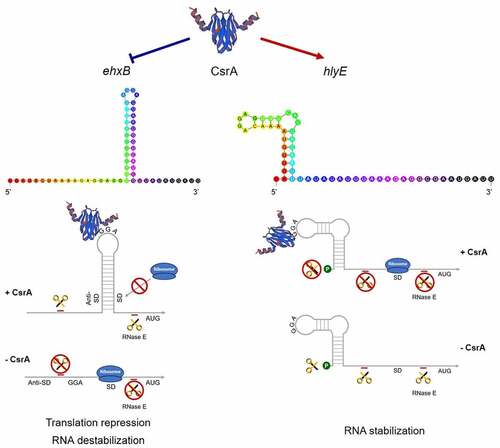
Figure 7. CsrA dual regulation of EHEC hemolysis.
Supplemental Material
Download Zip (66.9 KB)Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
