Figures & data
Figure 1. Multi-Protein and small peptide bacterial genotoxins. (a) Schematic diagram depicting the intoxication processes of multi-protein and small peptide bacterial genotoxins. (b) CDT structure. CdtB is the enzymatic ‘A’ subunit possessing DNase I-like activity. CdtA and CdtC are receptor-binding ‘B’ subunits of CDTs. (c) Typhoid toxin structure. CdtB is one of the ‘A’ subunits of typhoid toxin, possessing DNase I-like activity. PltA is the other ‘A’ subunit, while PltB forms the homopentameric receptor-binding ‘B’ subunits of typhoid toxin. (d) Representative colibactin structures. According to colibactin nomenclature, the numbers behind the word colibactin generally match their molecular weight [Citation94]. (b and c) panels are prepared using PyMOL. AaCDT, Aggregatibacter actinomycetemcomitans CDT (PDB: 2F2F). StyCdtb, Salmonella enterica subsp. enterica serovar Typhi CdtB (PDB: 4K6L). Created with BioRender.Com.
![Figure 1. Multi-Protein and small peptide bacterial genotoxins. (a) Schematic diagram depicting the intoxication processes of multi-protein and small peptide bacterial genotoxins. (b) CDT structure. CdtB is the enzymatic ‘A’ subunit possessing DNase I-like activity. CdtA and CdtC are receptor-binding ‘B’ subunits of CDTs. (c) Typhoid toxin structure. CdtB is one of the ‘A’ subunits of typhoid toxin, possessing DNase I-like activity. PltA is the other ‘A’ subunit, while PltB forms the homopentameric receptor-binding ‘B’ subunits of typhoid toxin. (d) Representative colibactin structures. According to colibactin nomenclature, the numbers behind the word colibactin generally match their molecular weight [Citation94]. (b and c) panels are prepared using PyMOL. AaCDT, Aggregatibacter actinomycetemcomitans CDT (PDB: 2F2F). StyCdtb, Salmonella enterica subsp. enterica serovar Typhi CdtB (PDB: 4K6L). Created with BioRender.Com.](/cms/asset/1a5579e5-6914-45ec-8a93-5f0861469867/kvir_a_2097417_f0001_oc.jpg)
Figure 2. CDT production, assembly, and trafficking. (a) The cdtA, cdtB, and cdtC genes are encoded within the same operon and expressed in the cytoplasm. ①, CdtA, CdtB, and CdtC are transported to the periplasm compartment (PC) through the Sec secretion pathway. Unlike CdtB and CdtC, CdtA is lipidated in the inner membrane (IM). ②, The lipoCdtA was transported onto the outer membrane (OM). ③, LipoCdtA on the OM recruits CdtC in the PC to form a heterodimer. ④, CdtC-lipoCdtA recruits a protease on the OM, resulting in the CdtA/CdtC heterodimer complex. ⑤, CdtA/CdtC heterodimer recruits free CdtB in the PC and forms CDT holotoxin. (b) Schematic diagram depicting the expression, assembly, trafficking, and intoxication processes of CDT: ①, CdtB/CdtC may be dissociated from CdtA in the endosome. ②, CdtB/CdtC transport to the ER through the retrograde transport pathway. ③, CdtB is dissociated from CdtC in the ER. ④, CdtB leaves the ER to the cytoplasm through an ERAD or ERAD-related pathway. ⑤, CdtB utilizes an atypical NLS sequence(s) to enter the nucleus and plays its function as a genotoxin. Created with BioRender.com.
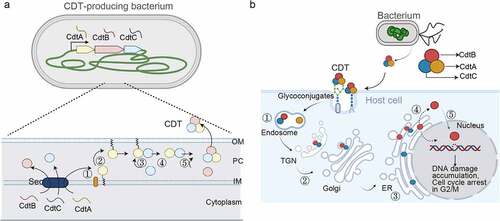
Figure 3. Typhoid toxin – two toxins become one. The gene organizations and 3D structures of pertussis toxin (a), AaCDT (b), and typhoid toxin (c). The enzymatic ‘A’ subunits and the receptor-binding ‘B’ subunits are indicated. Figures are prepared using PyMOL. Pertussis toxin (PDB: 1PRT). AaCDT, Aggregatibacter actinomycetemcomitans CDT (PDB: 2F2F). StyCdtB, Salmonella enterica serovar Typhi CdtB (PDB: 4K6 L).
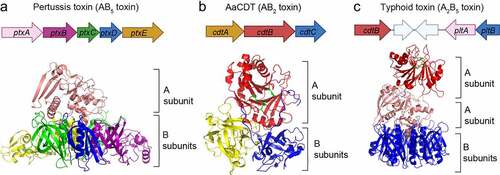
Figure 4. Typhoid toxin production, assembly, trafficking, and intoxication. (a) Typhoid toxin production, assembly, and secretion from S. Typhi. cdtB is transcribed from one operon, while pltB, pltA (and one additional gene, ttsA) are co-transcribed by another operon. ①, CdtB, PltA, PltB remains unstructured in the bacterial cytoplasm until they are transported into the PC through the Sec secretion pathway. The secreted CdtB, PltA, and PltB are self-assembled into typhoid toxin holotoxin. ②, Typhoid toxin enters the outer periplasm space by the action of the Tsa enzyme that breaks the peptidoglycan. ③, Typhoid toxin finally secretes to the lumen of the SCV. (b) Schematic diagram depicting typhoid toxin expression, assembly, trafficking, and intoxication. The steps involved are: ①, Salmonella invades host cells and forms the Salmonella-containing vacuole (SCV). ②, Salmonella in the SCV expresses and secretes the typhoid toxin under a specific SPI2-T3SS vacuole condition. The secreted typhoid toxin in the SCV binds to Neu5Ac-terminated glycoprotein receptors on the SCV and is sorted into anterograde vesicles. ③, the exocytosis of vesicles carrying typhoid toxin relies on the microtubule-based transport. Finally, typhoid toxin from S. Typhi-infected cells is released into the extracellular environment. ④, Typhoid toxin in the extracellular environment enters the target host cells through receptor-mediated endocytosis. Typhoid toxin utilizes multiantennary N-linked glycan receptors terminated in Neu5Ac. ⑤, the endocytosed typhoid toxin utilizes the retrograde transport pathway to enter the ER. ⑥, Typhoid toxin is disassembled into CdtB and PltA/pltB by a reductase. ⑦, PltA utilizes the ERAD pathway to translocate to the cytoplasm where it plays its ADP-ribosylation enzyme function. ⑧, CdtB utilizes an ERAD or ERAD-related pathway to leave the ER and ultimately be transported to the nucleus, where CdtB plays its genotoxic activity. Created with BioRender.Com.
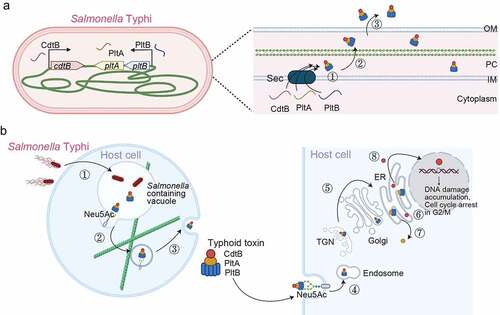
Figure 5. Colibactin intoxication processes. (a) a diagram depicting the biogenesis of colibactin. Colibactins are peptide-polyketide metabolites synthesized by enzymes encoded in the colibactin (Clb) gene cluster. Through a coordinated action by Clb enzymes, precolibactin is synthesized in the bacterial cytoplasm. The lower panel indicates key steps in the synthesis of colibactin in the periplasm: ①, Precolibactins synthesized in the bacterial cytoplasm are inactive, and are transported into the periplasm through the ClbM transporter. ②, ClbP in the periplasm cuts the prodrug motif of precolibactin, resulting in the active colibactin. ③, Colibactins are secreted from the bacteria through a direct membrane diffusion or yet-to-be-identified mechanism. (b) Schematic diagram depicting an intoxication process of colibactin. It is still unclear, but two mechanisms are proposed: (1) colibactin produced extracellularly is diffused to enter host cells and (2) intracellular bacteria produce colibactin in the host cell cytoplasm. Created with BioRender.Com.
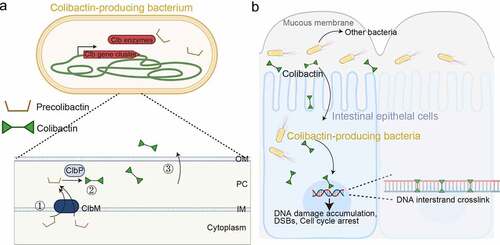
Figure 6. Colibactin biogenesis. The N-myristoyl-D-Asn prodrug side chains in the Precolibactin are indicated in orange, which is later removed by ClbP in the periplasm. Once the N-myristoyl-D-Asn prodrug side chain is removed, precolibactin is four-fold cyclized to form the colibactin structure. The unstable α-aminoketone group in precolibactin is indicated in red, which is oxidated and hydrolysed. The oxidated and hydrolysed form in the colibactin structure is indicated in green. The α-diketone group found in colibactin is also unstable and easily cleaved under a mildly nucleophilic condition. The electrophilic cyclopropane groups and the DNA alkylation reaction groups are labelled in dark blue and light blue, respectively. The macrocyclic and 5-hydroxy-oxazole groups are shown in the structure of colibactin 645.
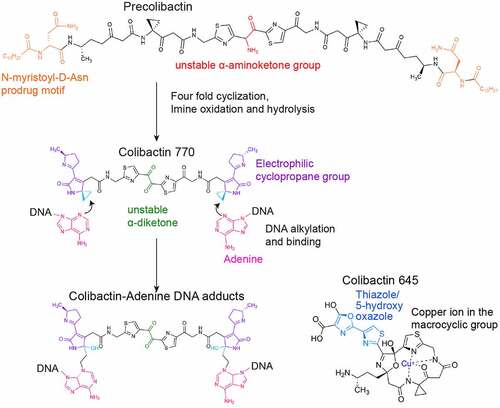
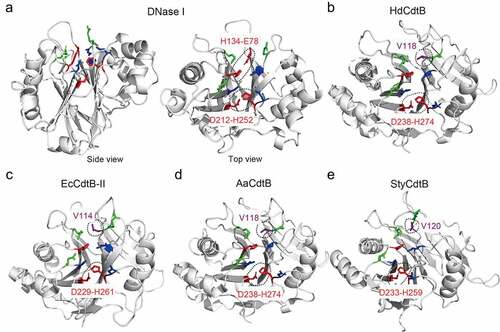
Figure 7. Key residues of DNaseI, HdCdtB, EcCdtB, AaCdtB, and StyCdtB. Structures of bovine DNase I (PDB: 2DNJ) (a), HdCdtb (PDB: 1SR4) (b), EcCdtb-II (PDB: 2F1N) (c), AaCdtb (PDB: 2F2F) (d), and StyCdtb (PDB: 4K6 L) (e). Catalytic (red), metal-binding (blue), and DNA binding (green) residues in their active site are indicated. Dotted circles are the catalytic histidine-partner amino acid hydrogen bonds, as well as the counterpart amino acid residues comparable to DNase I E78 that forms a hydrogen bond with H134. HdCdtb, Haemophilus ducreyi CdtB. EcCdtb. Escherichia coli CdtB. AaCdtb, Aggregatibacter actinomycetemcomitans. StyCdtb, Salmonella enterica subsp. enterica serovar Typhi CdtB. See also Supplemental . Figures are prepared using PyMOL. Bovine DNase I was used because of its structure complexed with DNA (PDB: 2DNJ).
Supplemental Material
Download MS Word (24.3 KB)Data availability statement
Data that support this review article are available upon request.
