Figures & data
Figure 1. CRP-Dependent activation of the EAEC 042 pic promoter. (a) the panel shows the organisation of the pic locus on the EAEC 042 chromosome, detailing the flanking IS110 and IS66 family elements. The exploded view shows a schematic representation of the picp042 promoter fragment. The CRP binding site is shown as inverted arrows and the start site of transcription (+1) is indicated by a bent arrow. The flanking EcoRI and HindIII sites used to clone the fragment are indicated. (b) the base sequence of the picp042 promoter fragment from EAEC 042. The CRP-binding site and two proposed −10 promoter elements are underlined and matches to their respective consensus sequences are in bold [Citation23,Citation24]. The transcription start site (+1) is lower case and indicated by a bent arrow and substitutions, which change the CRP-binding site and adjacent sequences, are shown. The bases that were identified by potassium permanganate footprinting as being single stranded in the open complex are highlighted in red. Terminal EcoRI and HindIII sites are bold and underlined. (c) the panel details β-galactosidase activities determined in the Δlac E. coli K-12 strain M182 and its δcrp derivative. Cells carried the lac expression vector pRW224 into which EAEC 042 picp042 promoter fragments were cloned. The p58A and p56C substitutions disrupt the CRP binding site at the pic promoter, see (b). Cells were cultured in LB medium and βgalactosidase activities are stated as nmol of ONPG hydrolysed min−1 mg−1 dry cell mass, activities are the average of at least three independent biological replicates. Standard deviations are shown and * indicates P < 0.01 using a Student’s t-test. (d) the panel shows the sequence trace from a 5` RACE experiment, which determined the EAEC 042 pic promoter transcription start site (TSS: green box). (e) End-labelled picp042 AatII-HindIII fragment was incubated with RNA polymerase and CRP and subjected to potassium permanganate footprinting. The concentration of CRP was as follows: lanes 1 and 2, no protein; lane 3, 400 nM; lane 4, 800 nM. Each reaction contained 50 nM RNA polymerase and 200 µm cAMP. Maxam-Gilbert ‘G+A’ sequencing reactions have been run to calibrate the gel. The location of cleavage sites within the EAEC 042 pic promoter are highlighted by stars (and in red in panel (b)).
![Figure 1. CRP-Dependent activation of the EAEC 042 pic promoter. (a) the panel shows the organisation of the pic locus on the EAEC 042 chromosome, detailing the flanking IS110 and IS66 family elements. The exploded view shows a schematic representation of the picp042 promoter fragment. The CRP binding site is shown as inverted arrows and the start site of transcription (+1) is indicated by a bent arrow. The flanking EcoRI and HindIII sites used to clone the fragment are indicated. (b) the base sequence of the picp042 promoter fragment from EAEC 042. The CRP-binding site and two proposed −10 promoter elements are underlined and matches to their respective consensus sequences are in bold [Citation23,Citation24]. The transcription start site (+1) is lower case and indicated by a bent arrow and substitutions, which change the CRP-binding site and adjacent sequences, are shown. The bases that were identified by potassium permanganate footprinting as being single stranded in the open complex are highlighted in red. Terminal EcoRI and HindIII sites are bold and underlined. (c) the panel details β-galactosidase activities determined in the Δlac E. coli K-12 strain M182 and its δcrp derivative. Cells carried the lac expression vector pRW224 into which EAEC 042 picp042 promoter fragments were cloned. The p58A and p56C substitutions disrupt the CRP binding site at the pic promoter, see (b). Cells were cultured in LB medium and βgalactosidase activities are stated as nmol of ONPG hydrolysed min−1 mg−1 dry cell mass, activities are the average of at least three independent biological replicates. Standard deviations are shown and * indicates P < 0.01 using a Student’s t-test. (d) the panel shows the sequence trace from a 5` RACE experiment, which determined the EAEC 042 pic promoter transcription start site (TSS: green box). (e) End-labelled picp042 AatII-HindIII fragment was incubated with RNA polymerase and CRP and subjected to potassium permanganate footprinting. The concentration of CRP was as follows: lanes 1 and 2, no protein; lane 3, 400 nM; lane 4, 800 nM. Each reaction contained 50 nM RNA polymerase and 200 µm cAMP. Maxam-Gilbert ‘G+A’ sequencing reactions have been run to calibrate the gel. The location of cleavage sites within the EAEC 042 pic promoter are highlighted by stars (and in red in panel (b)).](/cms/asset/1155a3e1-8e45-4dfc-90f6-5a961b66bbd6/kvir_a_2111754_f0001_oc.jpg)
Figure 2. CRP-Dependent activation of the UPEC CFT073 picU promoter. (a) the panel shows an alignment of the UPEC picp073 promoter sequence with the sequence from EAEC picp042. The CRP-binding site and −10 promoter elements are underlined, with matches to their respective consensus sequences in bold [Citation23,Citation24]. For both fragments, transcription start sites (+1) are lower case bold, translation initiation codons (GTG) are boxed and the Shine–Dalgarno sequences (SD) are bold and underlined. The position of the p37A and p34T substitutions, which disrupt the CRP-binding site in the UPEC picp073 promoter fragment, are shown and the bases, identified by potassium permanganate footprinting as being single stranded in the open complex, are in red. (b) the panel details β-galactosidase activities determined in the Δlac E. coli K-12 strain M182 and its δcrp derivative. Cells carried the lac expression vector pRW224 into which the UPEC picp073 promoter fragment was cloned. (c) the panel desplays β-galactosidase activities determined in the strain M182, with cells carrying various UPEC picp073 promoter fragments, cloned into pRW224. The p37A and p34T substitutions disrupt the CRP binding site within the pic073 promoter fragment, see (a). In both (b) and (c), cells were cultured in LB medium and β-galactosidase activities are the average of at least three independent biological replicates. Standard deviations are shown and * indicates P < 0.01 using a Student’s t-test. (d) the panel shows the sequence trace from a 5` RACE experiment, which determined the UPEC picU promoter transcription start site (TSS: green box). (e) End-labelled picp073 AatII-HindIII fragment was incubated with RNA polymerase and CRP and subjected to potassium permanganate footprinting. The concentration of CRP was as follows: lanes 1 and 2, no protein; lane 3, 400 nM; lane 4, 800 nM. Reactions contained 50 nM RNA polymerase and 200 µm cAMP and Maxam-Gilbert ‘G+A’ sequencing reactions have been included. The location of potassium permanganate cleavage sites are shown starred.
![Figure 2. CRP-Dependent activation of the UPEC CFT073 picU promoter. (a) the panel shows an alignment of the UPEC picp073 promoter sequence with the sequence from EAEC picp042. The CRP-binding site and −10 promoter elements are underlined, with matches to their respective consensus sequences in bold [Citation23,Citation24]. For both fragments, transcription start sites (+1) are lower case bold, translation initiation codons (GTG) are boxed and the Shine–Dalgarno sequences (SD) are bold and underlined. The position of the p37A and p34T substitutions, which disrupt the CRP-binding site in the UPEC picp073 promoter fragment, are shown and the bases, identified by potassium permanganate footprinting as being single stranded in the open complex, are in red. (b) the panel details β-galactosidase activities determined in the Δlac E. coli K-12 strain M182 and its δcrp derivative. Cells carried the lac expression vector pRW224 into which the UPEC picp073 promoter fragment was cloned. (c) the panel desplays β-galactosidase activities determined in the strain M182, with cells carrying various UPEC picp073 promoter fragments, cloned into pRW224. The p37A and p34T substitutions disrupt the CRP binding site within the pic073 promoter fragment, see (a). In both (b) and (c), cells were cultured in LB medium and β-galactosidase activities are the average of at least three independent biological replicates. Standard deviations are shown and * indicates P < 0.01 using a Student’s t-test. (d) the panel shows the sequence trace from a 5` RACE experiment, which determined the UPEC picU promoter transcription start site (TSS: green box). (e) End-labelled picp073 AatII-HindIII fragment was incubated with RNA polymerase and CRP and subjected to potassium permanganate footprinting. The concentration of CRP was as follows: lanes 1 and 2, no protein; lane 3, 400 nM; lane 4, 800 nM. Reactions contained 50 nM RNA polymerase and 200 µm cAMP and Maxam-Gilbert ‘G+A’ sequencing reactions have been included. The location of potassium permanganate cleavage sites are shown starred.](/cms/asset/3c4a609c-7fe3-4aab-9d5e-6f9215292f40/kvir_a_2111754_f0002_oc.jpg)
Figure 3. The EAEC and UPEC pic promoters use different −10 elements. (a) the panel details β-galactosidase activities from E. coli strain M182 Δcrp, carrying the lac expression vector pRW224 into which the EAEC picp042 or UPEC picp073 promoter fragments were cloned. Cells also carried either plasmid pDCRP or derivatives of pDCRP, encoding substitutions in CRP activating regions AR1 (HL159) and/or AR2 (KE101). (b) the panel shows schematic representations of the wild type EAEC picp042 and UPEC picp073 promoter fragments and fragments carrying substitutions in putative upstream and downstream −10 promoter elements. CRP binding sites are shown as inverted arrows and wild-type and mutant −10 elements are shown as filled or empty boxes, respectively. (c) the panel displays β-galactosidase activities in E. coli strain M182, carrying pRW224 into which the EAEC picp042 or UPEC picp073 promoter fragments depicted in (b) were cloned. Note that the numbering of promoter inserts is the same in both (b) and (c). For all experiments, cells were grown in LB, βgalactosidase activities are stated as nmol of ONPG hydrolysed min−1 mg−1 dry cell mass and are the average of at least three independent biological replicates. Standard deviations are shown and * indicates P < 0.01, ** indicates P < 0.05 and ns (not significant) P > 0.05, using a Student’s t-test.
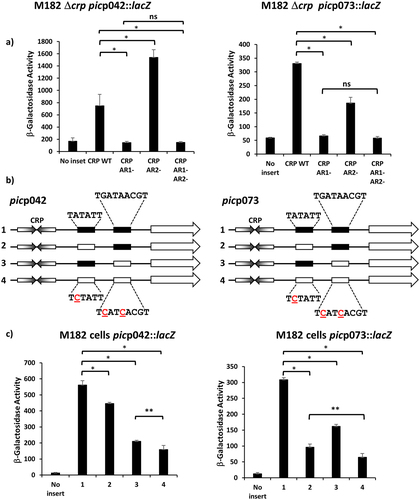
Figure 4. Translation efficiency differs for the EAEC picp042 and UPEC picp073 promoter fragments. (a) the panel shows schematic representations of the wild-type EAEC picp042 and UPEC picp073 promoter fragments and derivatives with substitutions in the upstream or downstream −10 promoter elements. CRP binding sites are shown as inverted arrows and wild-type and mutant −10 elements are shown as filled or empty white boxes, respectively. Fragments were cloned into pRW224 and pRW225 to generate lacZ transcription fusions and translation fusions, respectively. β-galactosidase activities were measured in E. coli M182 carrying either pRW224 or pRW225 into which the various EAEC picp042 and UPEC picp073 promoter fragments were cloned. Translation Efficiency is shown as the ratio of the values observed for pRW224 (transcription only) and pRW225 (transcription and translation) and is set as 1 for each starting wild type promoter. Cells were cultured in LB medium, βgalactosidase activities are the average of at least three independent values. (b) the panel shows the predicted secondary structure of the 5` ends of the mRNA when initiated from either the upstream or downstream -10 element in the UPEC picp073 fragment. The location of the downstream −10 element, the Shine-Dalgarno sequence (SD) and translation initiation codon (GUG) are shown.
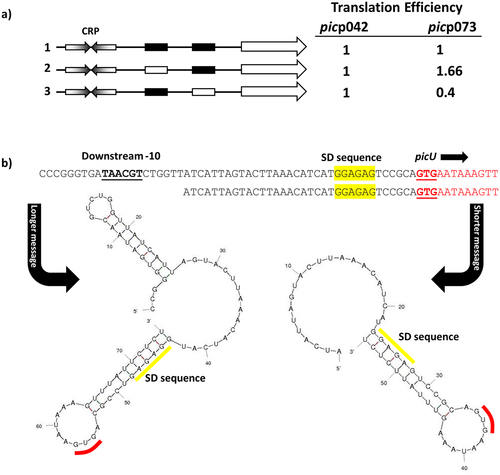
Figure 5. Fis represses the EAEC and UPEC pic promoters. (a) the panel illustrates β-galactosidase activities measured in the Δlac E. coli K-12 strain BW25113 and its δfis derivative. Cells carried the lac expression vector pRW224 into which the EAEC 042 picp042 or UPEC CFT073 picp073 promoter fragments were cloned. Cells were grown in LB medium and βgalactosidase activities are the average of at least three independent biological replicates. Standard deviations are shown and * indicates P < 0.01 using a student’s t-test. (b) the panel shows an SDS-PAGE gel run to quantify the amount of Pic protein secreted into the external medium by wild-type strain BW25113 (WT) and its Δfis and Δcrp derivatives. Where indicated, cells carried plasmid pPic, in which the EAEC 042 pic promoter and gene are cloned into plasmid pACYC184 [Citation27]. The arrows indicate the location of secreted Pic protein (116 kDa). PageRuler prestained protein ladder (Fisher Scientific) were run to calibrated the gel. (c) Gel retardation assays of the EAEC pic042 and UPEC pic073 promoter fragments with purified Fis protein. End-labelled pic promoter fragments were incubated with purified Fis protein: lanes 1–5, picp042 EcoRI-HindIII fragment; lanes, 6–10, picp073 EcoRI-HindIII fragment. The amount of Fis protein in each reaction was: lanes 1 and 6, no protein; lanes 2 and 7, 200 nM; lanes 3 and 8, 400 nM; lanes 4 and 9, 800 nM; lanes 5 and 10, 1.35 µm. The location of Fis/DNA complexes and unbound DNA is indicated.
![Figure 5. Fis represses the EAEC and UPEC pic promoters. (a) the panel illustrates β-galactosidase activities measured in the Δlac E. coli K-12 strain BW25113 and its δfis derivative. Cells carried the lac expression vector pRW224 into which the EAEC 042 picp042 or UPEC CFT073 picp073 promoter fragments were cloned. Cells were grown in LB medium and βgalactosidase activities are the average of at least three independent biological replicates. Standard deviations are shown and * indicates P < 0.01 using a student’s t-test. (b) the panel shows an SDS-PAGE gel run to quantify the amount of Pic protein secreted into the external medium by wild-type strain BW25113 (WT) and its Δfis and Δcrp derivatives. Where indicated, cells carried plasmid pPic, in which the EAEC 042 pic promoter and gene are cloned into plasmid pACYC184 [Citation27]. The arrows indicate the location of secreted Pic protein (116 kDa). PageRuler prestained protein ladder (Fisher Scientific) were run to calibrated the gel. (c) Gel retardation assays of the EAEC pic042 and UPEC pic073 promoter fragments with purified Fis protein. End-labelled pic promoter fragments were incubated with purified Fis protein: lanes 1–5, picp042 EcoRI-HindIII fragment; lanes, 6–10, picp073 EcoRI-HindIII fragment. The amount of Fis protein in each reaction was: lanes 1 and 6, no protein; lanes 2 and 7, 200 nM; lanes 3 and 8, 400 nM; lanes 4 and 9, 800 nM; lanes 5 and 10, 1.35 µm. The location of Fis/DNA complexes and unbound DNA is indicated.](/cms/asset/08c85161-a26e-4c21-b3df-b77f63494719/kvir_a_2111754_f0005_oc.jpg)
Supplemental Material
Download MS Word (216 KB)Data availability statement
All data relating to this article are present in the article and the accompanying Supplementary Material.
