Figures & data
Figure 1. Z-Score-Normalized haemocyte counts at 30 (a), 90 (b), and 180 (c) minutes post-injection of several norepinephrine (NE) concentrations in G. mellonella larvae. Box plots represent z-normalized haemocyte counts for n = 20 larvae per concentration. NE decreases the number of circulating haemocytes at 1 µM (Kruskal–Wallis test, p < 0.0001) after 30 minutes. * Significantly different (p < 0.05) compared with the phosphate-buffered saline (PBS) group.
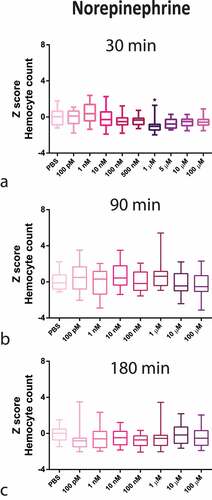
Figure 2. Z-Score-Normalized haemocyte counts at 30 (a), 90 (b), and 180 (c) minutes post-injection of a wide concentration range of isoproterenol (ISO) in G. mellonella larvae. Box plots represent z-normalized haemocyte counts for n = 20 larvae per concentration. ISO increases the number of haemocytes at 100 pM and 500 pM (Kruskal–Wallis test, p < 0.0001) after 30 min. * Significantly different (p < 0.05) compared with the phosphate-buffered saline (PBS) group.
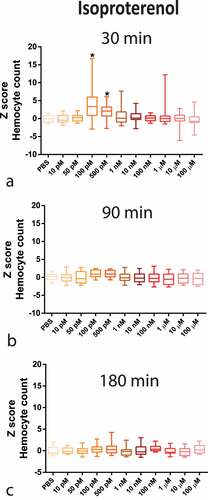
Figure 3. (a) Survival curve over 5 days of concomitant injection of 1 µM NE and 108 CFU/mL P. gingivalis (n = 20 larvae per group, Kaplan–Meier plot, Log-rank [Mantel-cox] p = 0.0109). (b) Survival curve after concomitant administration of 100 pM ISO and P. gingivalis (n = 20 larvae per group, Kaplan–Meier plot, log-rank [Mantel-cox] p = 0.0099). Same lower-case letters close to the graph lines/bars represent absence of significant differences.
![Figure 3. (a) Survival curve over 5 days of concomitant injection of 1 µM NE and 108 CFU/mL P. gingivalis (n = 20 larvae per group, Kaplan–Meier plot, Log-rank [Mantel-cox] p = 0.0109). (b) Survival curve after concomitant administration of 100 pM ISO and P. gingivalis (n = 20 larvae per group, Kaplan–Meier plot, log-rank [Mantel-cox] p = 0.0099). Same lower-case letters close to the graph lines/bars represent absence of significant differences.](/cms/asset/bdfdbd35-4302-47e8-9b8c-a0c198db6370/kvir_a_2123302_f0003_oc.jpg)
Figure 4. P. gingivalis recovery from the haemolymph of larvae infected with P. gingivalis ±1 µM NE (n = 10 pools of three larvae per group, mean and standard deviation, t-test, p = 0.1306) measured as CFU/mL (a) and P. gingivalis ±100 pM ISO (n = 10 pools of three larvae per group, mean and standard deviation, t-test, p = 0.0357) (b).
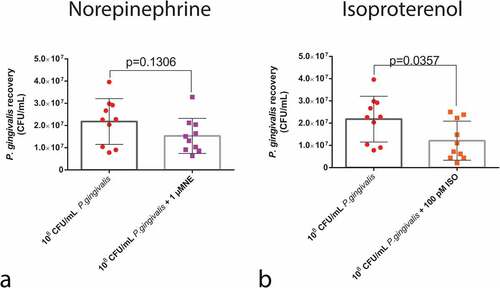
Figure 5. Humoral response of larvae to P. gingivalis infection. Colorimetric measurement of melanization for (a) NE (n = 20 larvae per group, Box plot, Kruskal–Wallis test, p < 0.0001) and (c) ISO (n = 20 larvae per group, Box plot, Kruskal–Wallis test, p < 0.0001). qPCR analysis of gloverin and cecropin mRNA expression 24 h after infection and NE administration (b) (gloverin: n = 8 larvae for PBS group and n = 10 larvae for P. gingivalis +/- 1 µM NE; Kruskal–Wallis test, p < 0.0001; cecropin : n = 10 larvae for PBS and P. gingivalis and n = 9 for P. gingivalis + 1 µM NE, Kruskal–Wallis test, p = 0.0024). (d) P. gingivalis +/- ISO (gloverin: n = 8 for PBS group, and n = 10 for P. gingivalis and P. gingivalis+ 100 pM ISO groups, Box plot, Kruskal–Wallis test, p < 0.0001; cecropin: n = 10 larvae per group, Kruskal–Wallis test, p = 0.0024). Same lower-case letters close to the graph lines/bars represent an absence of significant differences. * Significantly different compared with the PBS group.
Figure 6. Haemocyte count. Upper panels: Z-score-normalized haemocyte counts measured after P. gingivalis ±1 µM NE compared to PBS group (n = 20 larvae per group, mean and standard deviation, ANOVA, p < 0.0001) (a) and ±100pM ISO (n = 20 larvae per group, mean and standard deviation, ANOVA, p < 0.0001) (b). Same lower case letters close to the graph lines/bars represent the absence of significant differences. Bottom panels: representative image of an H&E-stained sagittal section of PBS or ISO (100 pM) injected larvae. Image shows the sessile haemocytes adhering to the fat body (FB), muscle (M), and trachea (T) (yellow arrowhead) and circulating haemocytes in the haemolymph (H) (orange arrowhead) (scale bar: 100 µm in the upper images and 40 µm in the insets). (c) Z-score-normalized haemocyte counts from haemolymph and fat body/adjacent organs in larvae that received PBS or 100pM ISO ± propranolol [10 µM] (n = 20 larvae per group, heatmap, ANOVA, p < 0.0001). * Significantly different compared with the PBS group (on haemolymph). Φ Significantly different compared with the PBS group (on fat body/adjacent organs).
![Figure 6. Haemocyte count. Upper panels: Z-score-normalized haemocyte counts measured after P. gingivalis ±1 µM NE compared to PBS group (n = 20 larvae per group, mean and standard deviation, ANOVA, p < 0.0001) (a) and ±100pM ISO (n = 20 larvae per group, mean and standard deviation, ANOVA, p < 0.0001) (b). Same lower case letters close to the graph lines/bars represent the absence of significant differences. Bottom panels: representative image of an H&E-stained sagittal section of PBS or ISO (100 pM) injected larvae. Image shows the sessile haemocytes adhering to the fat body (FB), muscle (M), and trachea (T) (yellow arrowhead) and circulating haemocytes in the haemolymph (H) (orange arrowhead) (scale bar: 100 µm in the upper images and 40 µm in the insets). (c) Z-score-normalized haemocyte counts from haemolymph and fat body/adjacent organs in larvae that received PBS or 100pM ISO ± propranolol [10 µM] (n = 20 larvae per group, heatmap, ANOVA, p < 0.0001). * Significantly different compared with the PBS group (on haemolymph). Φ Significantly different compared with the PBS group (on fat body/adjacent organs).](/cms/asset/8c41cc3a-3f9b-4ba4-aaef-c8513a9fcab8/kvir_a_2123302_f0006_oc.jpg)
Figure 7. Effects of adrenergic signalling on nodulation. (Top) Representative figures of larvae injected with P. gingivalis after 30 min, showing nodules formed around the fat body, muscles, and trachea (H&E staining). GI = gastrointestinal tract, FB = Fat body, M = muscle, T = trachea, H = haemolymph. (Bottom) No differences observed between P. gingivalis and P. gingivalis +1 µm NE regarding number of nodules counted on histological sections (n = 10 larvae per group, three sections per larvae, mean and standard deviation, t-test, p = 0.0851) (a). Decrease in the number of nodules in the P. gingivalis +100 pM ISO (n = 10 larvae per group, 3 sections per animal, mean and standard deviation, t-test, p = 0.0021) (b).
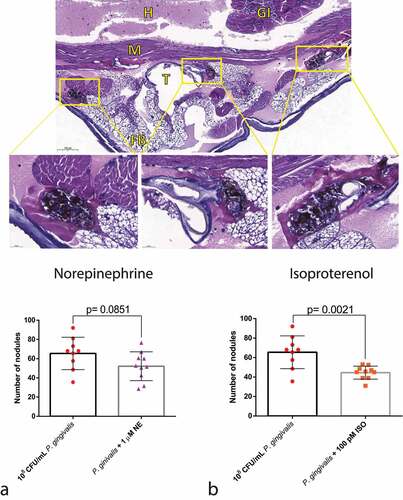
Figure 8. Direct action of adrenergic signalling on P. gingivalis. rgpb and fimA gene expression after co-culturing P. gingivalis ±1 µM or 100 µM NE (rgbp, n = 6 biological replicates per group, ANOVA, p < 0.0001; fimA, n = 6 biological replicates per group, mean and standard-deviation, ANOVA, p < 0.0001) (a). Co-culture of P. gingivalis with 1 µM or 100 µM NE had no effect on larvae survival 24 h after infection (n = 10 larvae for PBS group, n = 45 larvae for P. gingivalis group, and n = 20 larvae for PgNE 1 µM and PgNE 100 µM groups, Kaplan–Meier plot, log-rank [Mantel-cox] p = 0.0005) (b). Same lower-case letters close to the graph bars/lines represent an absence of significant difference.
![Figure 8. Direct action of adrenergic signalling on P. gingivalis. rgpb and fimA gene expression after co-culturing P. gingivalis ±1 µM or 100 µM NE (rgbp, n = 6 biological replicates per group, ANOVA, p < 0.0001; fimA, n = 6 biological replicates per group, mean and standard-deviation, ANOVA, p < 0.0001) (a). Co-culture of P. gingivalis with 1 µM or 100 µM NE had no effect on larvae survival 24 h after infection (n = 10 larvae for PBS group, n = 45 larvae for P. gingivalis group, and n = 20 larvae for PgNE 1 µM and PgNE 100 µM groups, Kaplan–Meier plot, log-rank [Mantel-cox] p = 0.0005) (b). Same lower-case letters close to the graph bars/lines represent an absence of significant difference.](/cms/asset/874d8dc5-a921-4b88-b554-74163e29cd7e/kvir_a_2123302_f0008_oc.jpg)
Figure 9. Direct action of β-adrenergic signalling on P. gingivalis. A) rgbp and fimA gene expression after co-culturing P. gingivalis ±100pM or 100 µM ISO (rgbp, n = 6 biological replicates per group, mean and standard deviation, ANOVA, p < 0.0001; fimA, n = 6 biological replicates per group, ANOVA, p = 0.0131) (a). Co-culturing P. gingivalis with 100 pM and 100 µM ISO increased the mortality of the larvae 24 h after infection (n = 10 larvae for PBS group, n = 45 larvae for P. gingivalis group, n = 20 for PgISO 100 pM and PgISO 100 µm and 100 pM ISO + PgISO 100 pM groups, Kaplan–Meier plot, log-rank [Mantel-cox] p < 0.0001) (b). The systemic administration of 100 pM ISO could partially reverse the increased PgISO 100pM phenotype (c). Same lower-case letters close to the graph bars/lines represent an absence of significant difference.
![Figure 9. Direct action of β-adrenergic signalling on P. gingivalis. A) rgbp and fimA gene expression after co-culturing P. gingivalis ±100pM or 100 µM ISO (rgbp, n = 6 biological replicates per group, mean and standard deviation, ANOVA, p < 0.0001; fimA, n = 6 biological replicates per group, ANOVA, p = 0.0131) (a). Co-culturing P. gingivalis with 100 pM and 100 µM ISO increased the mortality of the larvae 24 h after infection (n = 10 larvae for PBS group, n = 45 larvae for P. gingivalis group, n = 20 for PgISO 100 pM and PgISO 100 µm and 100 pM ISO + PgISO 100 pM groups, Kaplan–Meier plot, log-rank [Mantel-cox] p < 0.0001) (b). The systemic administration of 100 pM ISO could partially reverse the increased PgISO 100pM phenotype (c). Same lower-case letters close to the graph bars/lines represent an absence of significant difference.](/cms/asset/1ccdc8c5-1717-49e2-bc53-8d2455265ccf/kvir_a_2123302_f0009_oc.jpg)
Table 1. Genes investigating P. gingivalis virulence and G. mellonella immune defense.
Figure 10. Direct action of β-adrenergic signalling on P. gingivalis. A) rgbp and fimA gene expression after co-culturing P. gingivalis +100pM or 100 μM ISO (rgbp, n = 6 biological replicates per group, mean and standard deviation, ANOVA, p < 0.0001; fimA, n=6 biological replicates per group, ANOVA, p = 0.0131) (a). Co-culturing P. gingivalis with 100 pM and 100 μM ISO increased the mortality of the larvae 24 h after infection (n = 10 larvae for PBS group, n = 45 larvae for P. gingivalis group, n = 20 for PgISO 100 pM and PgISO 100 μM and 100 pM ISO + PgISO 100 pM groups, Kaplan–Meier plot, log-rank [Mantel-cox] p < 0.0001) (b). The systemic administration of 100 pM ISO could partially reverse the increased PgISO 100pM phenotype (c). Same lower-case letters close to the graph bars/lines represent an absence of significant difference.
![Figure 10. Direct action of β-adrenergic signalling on P. gingivalis. A) rgbp and fimA gene expression after co-culturing P. gingivalis +100pM or 100 μM ISO (rgbp, n = 6 biological replicates per group, mean and standard deviation, ANOVA, p < 0.0001; fimA, n=6 biological replicates per group, ANOVA, p = 0.0131) (a). Co-culturing P. gingivalis with 100 pM and 100 μM ISO increased the mortality of the larvae 24 h after infection (n = 10 larvae for PBS group, n = 45 larvae for P. gingivalis group, n = 20 for PgISO 100 pM and PgISO 100 μM and 100 pM ISO + PgISO 100 pM groups, Kaplan–Meier plot, log-rank [Mantel-cox] p < 0.0001) (b). The systemic administration of 100 pM ISO could partially reverse the increased PgISO 100pM phenotype (c). Same lower-case letters close to the graph bars/lines represent an absence of significant difference.](/cms/asset/d377e9ad-2732-4dc5-9717-49a9ca8cc6c6/kvir_a_2123302_f0010_oc.jpg)
