Figures & data
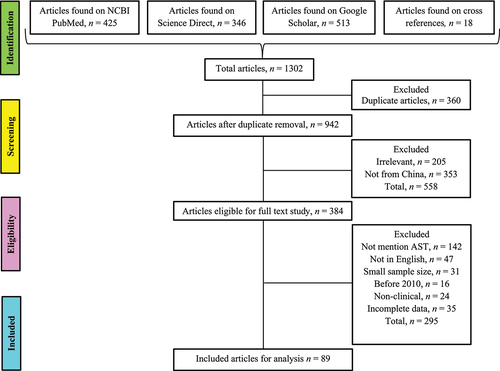
Table 1 Characteristics of articles include in current systematic analysis (n = 89)
Table 2 The molecular typing of Candida species mentioned in the included studies
Figure 2. Prevalence of Candida species, (a) the total occurrence of Candida species, the numerical on the top of the bar is the number of specific Candida species, (b) the occurrence of Candida species in different regions of China, each region is represented by specific colour as shown in the box, Multiple locations mean the that studies mentioned more than one region of China, (c) Prevalence of Candida species in association with various infection type, OC; oral candidiasis, UTI; urinary tract infection, NM; not mentioned the infection type, MI; multiple infections, involved in more than one infections, VVC; vulvovaginal candidiasis, BSI; bloodstream infection, IC; invasive candidiasis.
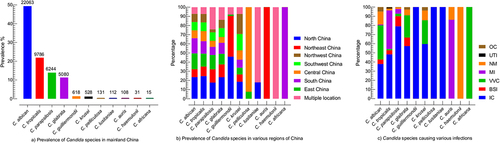
Figure 3. Antifungal susceptibility patterns of Candida albicans in the form of median susceptibility/wild type with 95% confidence interval.
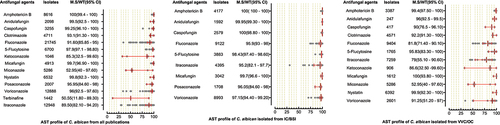
Figure 4. Antifungal susceptibility patterns of Candida tropicalis in the form of median susceptibility/wild type with 95% confidence interval.
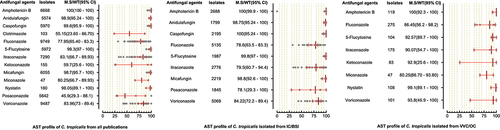
Figure 5. Antifungal susceptibility patterns of Candida parapsilosis in the form of median susceptibility/wild type with 95% confidence interval.
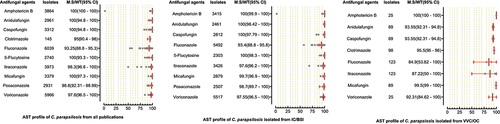
Figure 6. Antifungal susceptibility patterns of Candida glabrata in the form of median susceptibility/wild type with 95% confidence interval.
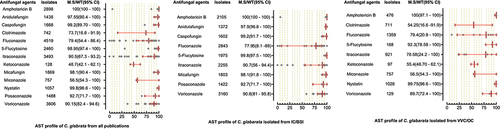
Figure 7. Antifungal susceptibility patterns of Candida krusei in the form of median susceptibility/wild type with 95% confidence interval.

Figure 8. Antifungal susceptibility patterns of C. auris, C. pelliculosa, C. guilliermondii and C. lusitaniae in the form of median susceptibility/wild type with 95% confidence interval.
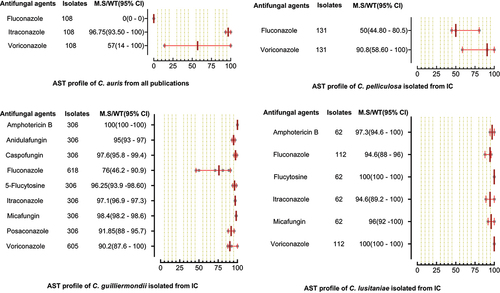
Table 3 Molecular mechanisms of antifungal drug resistance stated in the included studies
Supplemental Material
Download PDF (377.4 KB)Data availability statement
Data supporting our study can be provided by demand from the corresponding author (email: [email protected]) or the first author (email: [email protected]).