Figures & data
Table 1. Yield of GSL isolated from 14 pooled Atlantic salmon (20 g each).
Figure 1. Summary of neutral GSL found in Atlantic salmon epithelia. Symbol explanation: yellow square=N-acetylgalactosamine (GalNac), yellow circle=galactose (Gal), blue square=N-acetylglucosamine (GlcNac), blue circle=glucose (Glc), white square=N-acetylhexosamine (HexNac) and red triangle=fucose (Fuc).
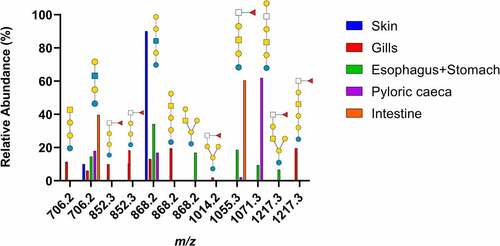
Figure 2. MS/MS interpretation of two fucosylated structures from Atlantic salmon gills. A) m/z 852 suggest a fucosylated globo structure. B) m/z 1217 suggest an elongated fucosylated globo structure with a 1–4 linkage between the terminal end hexose and middle HexNac giving the 0,2A4 cross ring fragment. Symbol explanation: yellow square=N-acetylgalactosamine (GalNac), yellow circle=galactose (Gal), blue circle=glucose (Glc), blue square=N-acetylglucosamine (GlcNac), white square=N-acetylhexosamine (HexNac) and red triangle=fucose (Fuc).
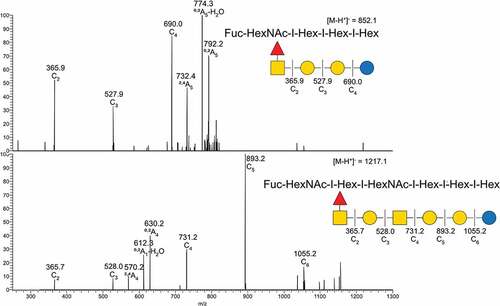
Figure 3. Summary of acid GSL found in Atlantic salmon epithelia. Symbol explanations: yellow square=N-acetylgalactosamine (GalNac), yellow circle=galactose (Gal), blue square=n-acetylglucosamine (GlcNac), blue circle=glucose (Glc), white square=N-acetylhexosamine (HexNac), red triangle=fucose (Fuc), purple diamond= N-acetylneuraminic acid (Neu5ac) and S=sulphate group.
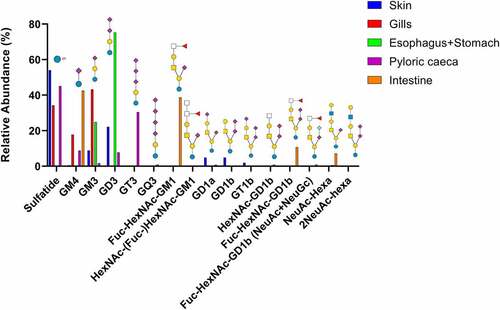
Figure 4. MS/MS of two native acid GSL from Atlantic salmon intestine. A) MS/MS of m/z 995.8 B) MS/MS of m/z 926.8. Symbol explanations: yellow square=N-acetylgalactosamine (GalNac), yellow circle=galactose (Gal), blue circle=glucose (Glc), red triangle=fucose (Fuc) and purple diamond= N-acetylneuraminic acid (Neu5ac).
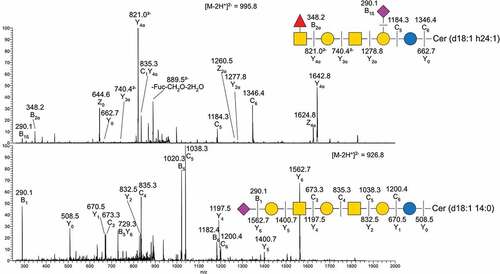
Figure 5. Distribution and A. salmonicida binding to α2–3 and α2–6 linked Neu5Ac. A) the relative abundance of α2–3 and α2–6 linked Neu5Ac on mucins and GSLs. Note that some glycan structures contain both α2–3 and α2–6 linked sialic acid. B) Binding of A. salmonicida to α2–3 and α2–6 sialyl-lacto HSA conjugates (8 µg/ml). No binding to α2–3 linked sialic acid was detected when compared to control (PBS), while the α2–6 linked sialic acids showed significant binding (n = 8). Data are shown after subtracting the average background control signal. Significance was calculated using Student’s-t test (****=p < 0.0001). Outliers in the background control (blank) were removed by ROUT (Q = 1%).
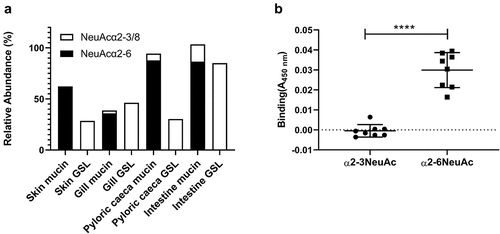
Figure 6. Comparison of terminal epitopes and size between mucins and GSLs. A, B, D, E, G, H, J and K) Comparison of terminal epitopes from GSL and mucin O-glycans. The acid and neutral GSL data were proportionally combined based on isolation data from table 1. The data was calculated based on the weight of the GSL (neutral/acid GSL) and number of terminal moieties. C, F, I and L) Comparison of number of monosaccharides in the GSL or O-glycans from the same organ tissue, based on previously published data [Citation8–11]. *neutral GSL containing one or two monosaccharides could not be included due to having too small mass for MS detection. # Mucin glycans containing a single monosaccharide can not be detected either.
![Figure 6. Comparison of terminal epitopes and size between mucins and GSLs. A, B, D, E, G, H, J and K) Comparison of terminal epitopes from GSL and mucin O-glycans. The acid and neutral GSL data were proportionally combined based on isolation data from table 1. The data was calculated based on the weight of the GSL (neutral/acid GSL) and number of terminal moieties. C, F, I and L) Comparison of number of monosaccharides in the GSL or O-glycans from the same organ tissue, based on previously published data [Citation8–11]. *neutral GSL containing one or two monosaccharides could not be included due to having too small mass for MS detection. # Mucin glycans containing a single monosaccharide can not be detected either.](/cms/asset/59bd3ec7-e082-449c-b0e6-b20a9eadb51a/kvir_a_2132056_f0006_oc.jpg)
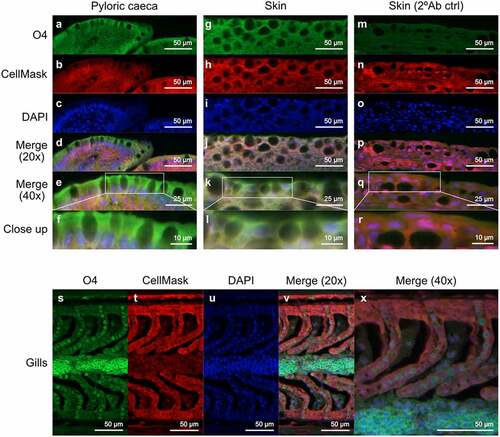
Figure 7. Location of sulfatide in Atlantic salmon pyloric caeca, skin and gills. (A, G and S) sulfatide tissue localization was visualized with the O4 antibody (green). (B, H, N and T) the tissue was outlined by CellMask (red). (C, I, O and U) DAPI (blue) stained the nuclei. (D, J, P and V) show the channels merged at 20x and (E, K, Q and X) at 40x. (F, L and R) Show close up images of the regions enclosed by white boxes in (E, K, Q and X). (M-R) show skin tissue without the O4 antibody, used as a background control.