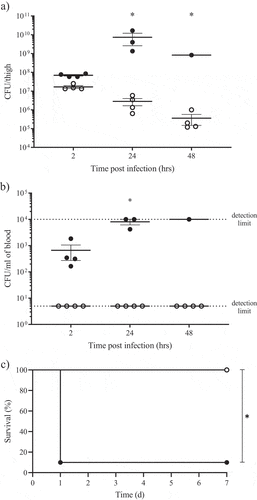
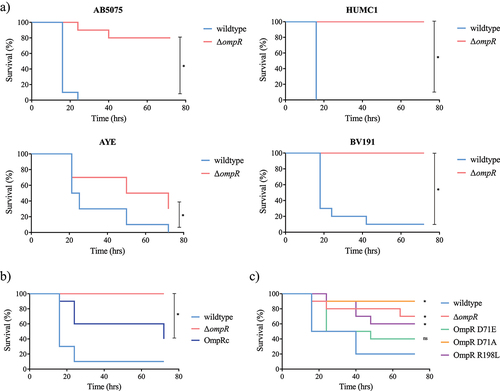
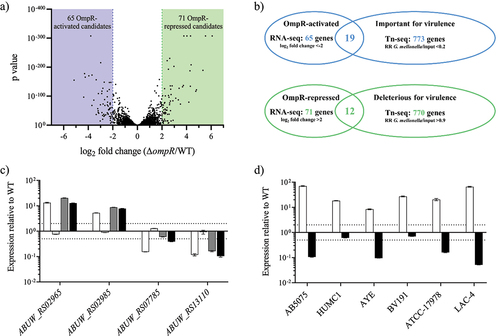
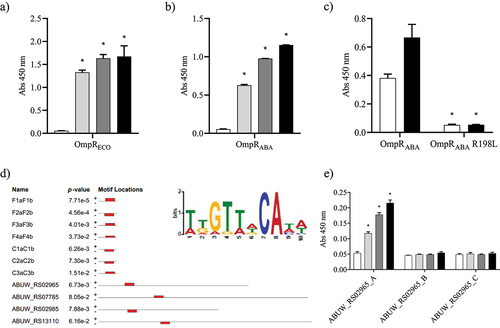
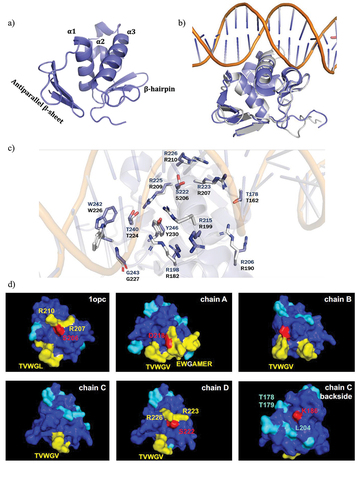
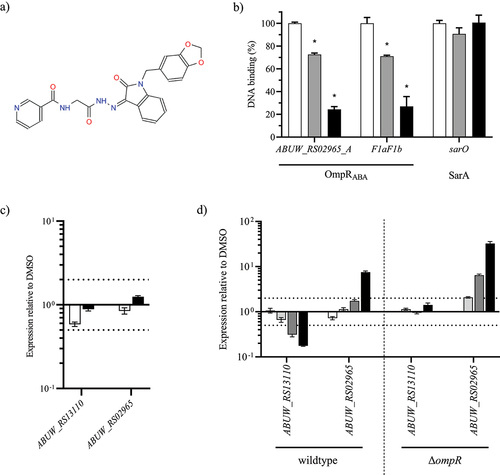
A) RNA-seq results representation of differentially expressed genes between AB5075 wildtype and its Δ
ompR mutants. Sixty-five OmpR-activated candidate genes (<-2 log
2 fold change, blue zone) and 71 OmpR-repressed candidate genes (>2 log
2 fold change, green zone) were identified. B) The RNA-seq results were combined with a Tn-seq dataset conducted to identify
A. baumannii genes that are important or deleterious for
A. baumannii virulence [
Citation26]. Nineteen OmpR-activated candidate genes were important for
A. baumannii virulence whereas 12 OmpR-repressed candidate genes were deleterious for
A. baumannii virulence. RR: read ratio. C) The transcript levels were determined in
A. baumannii AB5075 wildtype, AB5075 Δ
ompR (white), AB5075 OmpR D71E (light grey), AB5075 OmpR D71A (dark grey) and AB5075 OmpR R198L (black) mutants by quantitative real-time PCR and the expression level was normalized to the expression of the AB5075 wildtype strain (means ± SEM of two technical replicates). D) The transcript levels of
ABUW_RS02965 (white) and
ABUW_RS13110 (black) were determined in
A. baumannii Δ
ompR mutants and normalized to the transcript levels of their respective wildtype strains (means ± SEM of two technical replicates). Horizontal dotted lines depict a 2-fold up- or downregulation.