Figures & data
Figure 1. Parasitization reduced the total count of P. xylostella haemocytes via apoptosis. (a) the total haemocyte count (THC) of P. xylostella at 4 h pp by C. vestalis. (b) the survival ratio of P. xylostella haemocytes at 6 h, 2 h, and 4 h pp. Error bars indicate ± SD. Differences among samples were tested with tukey-test (ns: no significance; *** p<0.001). (c) TUNELlabeling to observe changes of P. xylostella apoptotic haemocytes at 4 h pp. Hemocytes were dissected and isolated, nuclei were stained with DAPI (blue), and apoptotic cells were labelled by TUNEL (red) (scale bar = 0 μm).
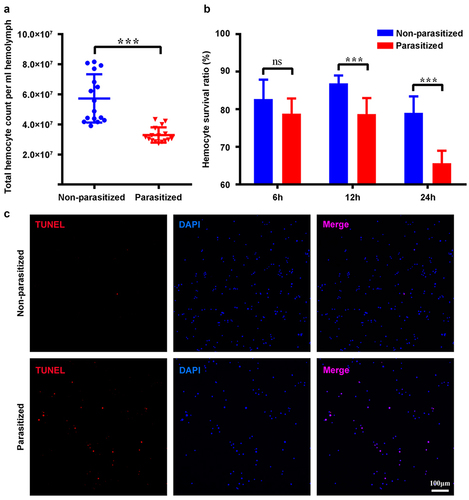
Figure 2. The activated CvBV induced apoptosis of host haemocytes. TUNEL apoptotic labelling to observe changes of apoptotic haemocytes of P. xylostella at 4 h post-injection of PBS, venom, inactivated and activated CvBV. The dosages of CvBV and venom were 0.05 FE. CvBV virions were put under UV-light for 2 hours to incapacitate the virus. Hemocytes were dissected and isolated, nuclei were stained with DAPI (blue), and apoptotic cells were labelled by TUNEL (red) (scale bar = 0 μm).
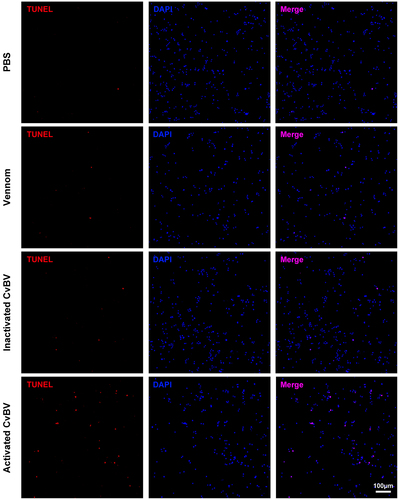
Figure 3. Expression levels of CvBV genes in P. xylostella haemocytes. CvBV gene expression levels in P. xylostella haemocytes at 6 h (a), 2 h (b), and 4 h (c) pp by C. vestalis. The transcripts were classified according to their location in the genome. 35 non-redundant circular CvBV genome was represented as linear molecules to visualize size and scale bar is in bps. For each circle, grey lines represent intergenic regions and different colours indicate the FPKMs of CvBV genes (see the list in S9, S10, and S11 Tables). Relative expression levels of CvBV genes in P. xylostella haemocytes were analysed using qPCR at 6 h (d), 2 h (e), and 4 h (f) pp. The right y-axis represents their FPKMs (green line) based on transcriptome data and the left ordinate represents relative expression levels (yellow columns). Error bars indicate ± SD.
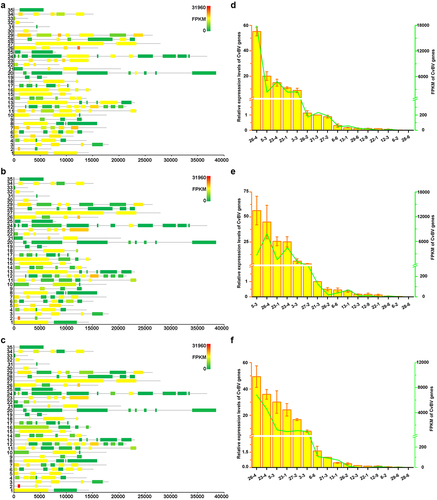
Figure 4. Overexpression of CvBV-26-4 induced apoptosis of P. xylostella haemocytes. The TUNEL apoptotic labelling to observe changes of haemocytes infected by modified baculoviruses (Bac-GFP, Bac-CvBV-2-1, Bac-CvBV-3-,5 and Bac-CvBV-26-4). Hemocytes were dissected and isolated, nuclei were stained with DAPI (blue), and apoptotic cells were labelled by TUNEL (red) (scale bar = 0 μm).
Figure 5. Expression profile of CvBV-26-4 in P. xylostella. (a) CvBV-26-4 expression profile in P. xylostella at 4 h, 8 h, 2 h, 6 h, and 20 h pp. (b) CvBV-26-4 expression profile in P. xylostella at 0.5 h, 1 h, 2 h, 4 h, 8 h, and 6 h pp. (c) the expression profile of CvBV-26-4 was measured by qPCR in 8 tissues of P. xylostella at 4 h pp including the epidermis, silk gland, fat body, midgut, haemocytes, central nervous system (CNS), malpighian tubules (MT), and testis. Error bars indicate ± SD (n = 3). Samples were compared with the tukey-test (**: indicates samples with a significant difference p<0.01).
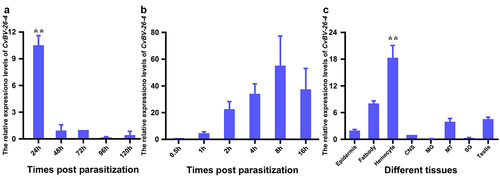
Figure 6. CvBV-26-4 is involved in the apoptosis of P. xylostella haemocytes. (a) the expression levels of CvBV-26-4 were measured by qPCR 4 h after the interference of CvBV-26-4 was introduced. Error bars indicate ± SD (n = 3). (b) Western blot of CvBV-26-4 in host haemolymph 4 h after the interference of CvBV-26-4. (c) TUNEL labelling of apoptotic haemocytes from P. xylostella at 4 h pi with rabbit IgG or CvBV-26-4 antiserum. Hemocytes were dissected and isolated, nuclei were stained with DAPI (blue), and apoptotic cells were labelled by TUNEL (red) (scale bar = 0 μm). (d) the percentage of apoptotic haemocytes from P. xylostella at 4 h post-injection of normal rabbit IgG or the CvBV-26-4 antiserum (n = 10). Differences among samples were tested with tukey-test (***: statistically significant p<0.001).
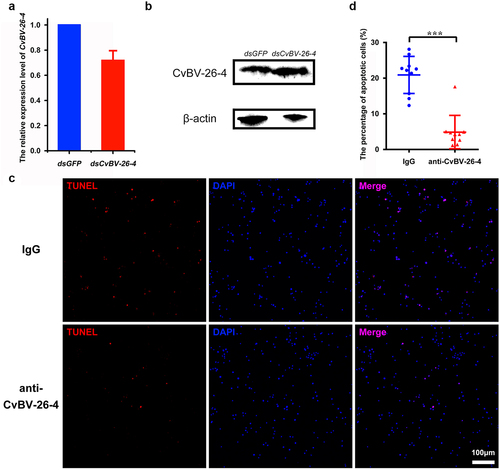
Figure 7. CvBV-26-4 homologs induced apoptosis of P. xylostella haemocytes. (a) amino acid sequence alignments of CvBV-26-4 and its homologs in Cotesia and glyptapanteles. The red box indicates the signal peptides. GYPY motifs conserved in CvBV-26-4 and its homologs are marked by red **** above the sequences. The numbers on the right are the position of the final amino acid. (b) the TUNEL apoptotic labelling of haemocytes at 4 h post-infection by modified baculoviruses (Bac-GFP, Bac-CcBV_CCQ71098.1, and Bac-CsKBV_CCQ19291.1). Hemocytes were dissected and isolated, nuclei were stained with DAPI (blue), and apoptotic cells were labelled by TUNEL (red) (scale bar = 0 μm).
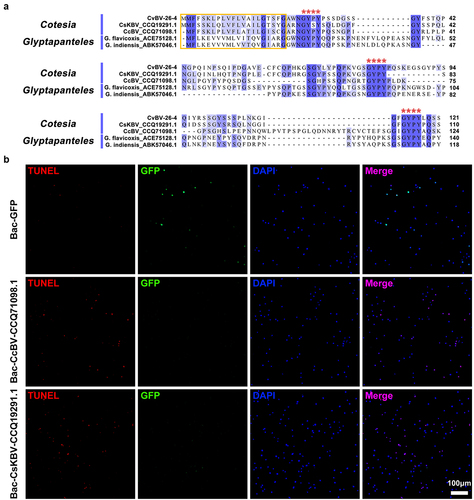
Figure 8. CvBV-26-4 peptide induced apoptosis of P. xylostella haemocytes. (a) amount of CvBV-26-4 protein in the haemocytes (top) and plasma (bottom) of P. xylostella at 6 h, 2 h, and 4 h pp. A band in the size range of 13 kDa that corresponds to the predicted CvBV-26-4 protein. Western blot are reported in supplementary information (figure S7). (b) the locations of CvBV 26–4 peptide (red) identified in the plasma of P. xylostella at 4 h pp by mass spectrometry. PSMs = 14. The conserved GYPY motifs were marked by underline. (c) the TUNEL apoptotic labelling of apoptotic haemocytes of P. xylostella at 4 h post-injection of the CvBV-26-4 peptide. (d) TUNEL labelling of apoptotic haemocytes in parasitized P. xylostella 4 h post-injection with CvBV-26-4 antibody and CvBV-26-4 peptide. Hemocytes were dissected and isolated, nuclei were stained with DAPI (blue), and apoptotic cells were labelled by TUNEL (red) (scale bar = 0 μm).
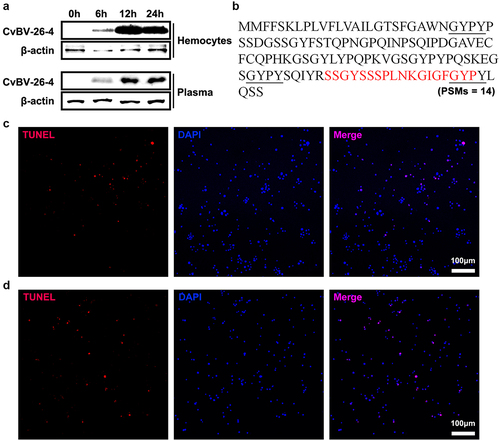
Figure 9. CvBV-26-4 induced apoptosis of D. melanogaster haemocytes. (a) morphologies of 3rd instar fly haemocytes that ectopically expressed CvBV-26-4 (scale bar = 0 μm). (b)apoptotic labelling of Drosophila larvae blood cells that ectopically expressed CvBV-26-4. Hemocytes were dissected and isolated, nuclei were stained with DAPI (blue), and apoptotic cells were labelled by TUNEL (red) (scale bar = 0 μm). (c) representative survival graphs of adult male Drosophila ectopically expressing CvBV-26-4 after injection of S. aureus (OD600 0.4). n = 20–25 flies. Experiments were performed in triplicate. (d) apoptotic staining of haemocytes from 3rd instar Drosophila larvae (W1118) at 4 h post-injection of 0.1 μg CvBV BAP peptide (injection of PBS was used as a negative control). Hemocytes were dissected out and nuclei were stained with DAPI (blue) and apoptotic cells were labelled by TUNEL (red) (scale bar = 0 μm).
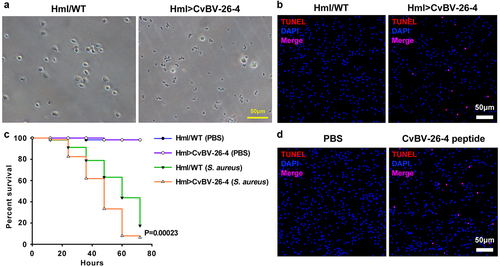
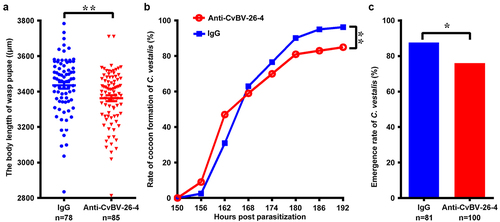
Figure 10. Interference of CvBV-26-4 on wasp development. 3rd instar P. xylostella larvae were injected with CvBV-26-4 antibody or rabbit IgG before parasitization. (a) body lengths of 1-day wasp pupae (µm). Error bars indicate ± SD. Differences among samples were tested with tukey-test (**: significant difference between samples p<0.01). Rate of wasp pupa formation (b) and eclosion rate of C. vestalis (c) in 3rd instar P. xylostella larvae injected with CvBV-26-4 antibody or rabbit IgG before parasitization. Differences among samples were tested with the chi-squared test. (*: significant difference between samples p <.
Supplemental Material
Download MS Word (1.7 MB)Data availability statement
The raw data for mRNA sequencing of P. xylostella haemocytes are deposited at SRA database of NCBI at https://www.ncbi.nlm.nih.gov, with accession number SRR10863199~SRR10863201.
